Modulation of receptor-like transmembrane kinase 1 nuclear localization by DA1 peptidases in Arabidopsis
- PMID: 36161927
- PMCID: PMC9546594
- DOI: 10.1073/pnas.2205757119
Modulation of receptor-like transmembrane kinase 1 nuclear localization by DA1 peptidases in Arabidopsis
Abstract
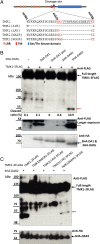
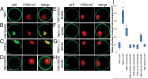
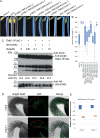
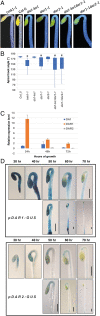
The cleavage of intracellular domains of receptor-like kinases (RLKs) has an important functional role in the transduction of signals from the cell surface to the nucleus in many organisms. However, the peptidases that catalyze protein cleavage during signal transduction remain poorly understood despite their crucial roles in diverse signaling processes. Here, we report in the flowering plant Arabidopsis thaliana that members of the DA1 family of ubiquitin-regulated Zn metallopeptidases cleave the cytoplasmic kinase domain of transmembrane kinase 1 (TMK1), releasing it for nuclear localization where it represses auxin-responsive cell growth during apical hook formation by phosphorylation and stabilization of the transcriptional repressors IAA32 and IAA34. Mutations in DA1 family members exhibited reduced apical hook formation, and DA1 family-mediated cleavage of TMK1 was promoted by auxin treatment. Expression of the DA1 family-generated intracellular kinase domain of TMK1 by an auxin-responsive promoter fully restored apical hook formation in a tmk1 mutant, establishing the function of DA1 family peptidase activities in TMK1-mediated differential cell growth and apical hook formation. DA1 family peptidase activity therefore modulates TMK1 kinase activity between a membrane location where it stimulates acid cell growth and initiates an auxin-dependent kinase cascade controlling cell proliferation in lateral roots and a nuclear localization where it represses auxin-mediated gene expression and growth.
Keywords: Arabidopsis; DA1 peptidase; TMK1 receptor kinase; auxin signaling; protein cleavage.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
-
- Adamiec M., Ciesielska M., Zalaś P., Luciński R., Arabidopsis thaliana intramembrane proteases. Acta Physiol. Plant. 39, 146 (2017).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

