Rpd3 regulates single-copy origins independently of the rDNA array by opposing Fkh1-mediated origin stimulation
- PMID: 36161938
- PMCID: PMC9546531
- DOI: 10.1073/pnas.2212134119
Rpd3 regulates single-copy origins independently of the rDNA array by opposing Fkh1-mediated origin stimulation
Abstract
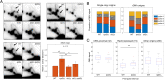
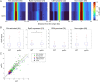
Eukaryotic chromosomes are organized into structural and functional domains with characteristic replication timings, which are thought to contribute to epigenetic programming and genome stability. Differential replication timing results from epigenetic mechanisms that positively and negatively regulate the competition for limiting replication initiation factors. Histone deacetylase Sir2 negatively regulates initiation of the multicopy (∼150) rDNA origins, while Rpd3 histone deacetylase negatively regulates firing of single-copy origins. However, Rpd3's effect on single-copy origins might derive indirectly from a positive function for Rpd3 in rDNA origin firing shifting the competitive balance. Our quantitative experiments support the idea that origins compete for limiting factors; however, our results show that Rpd3's effect on single-copy origin is independent of rDNA copy-number and of Sir2's effects on rDNA origin firing. Whereas RPD3 deletion and SIR2 deletion alter the early S phase dynamics of single-copy and rDNA origin firings in opposite fashion, unexpectedly only RPD3 deletion suppresses overall rDNA origin efficiency across S phase. Increased origin activation in rpd3Δ requires Fkh1/2, suggesting that Rpd3 opposes Fkh1/2-origin stimulation, which involves recruitment of Dbf4-dependent kinase (DDK). Indeed, Fkh1 binding increases at Rpd3-regulated origins in rpd3Δ cells in G1, supporting a mechanism whereby Rpd3 influences initiation timing of single-copy origins directly through modulation of Fkh1-origin binding. Genetic suppression of a DBF4 hypomorphic mutation by RPD3 deletion further supports the conclusion that Rpd3 impedes DDK recruitment by Fkh1, revealing a mechanism of Rpd3 in origin regulation.
Keywords: Chromatin domains; Genome instability; Histone deacetylase; Repetitive DNA; Replication origin.
Conflict of interest statement
The authors declare no competing interest.
Figures




References
-
- Tsang E., Carr A. M., Replication fork arrest, recombination and the maintenance of ribosomal DNA stability. DNA Repair (Amst.) 7, 1613–1623 (2008). - PubMed
-
- Nomura M., Ribosomal RNA genes, RNA polymerases, nucleolar structures, and synthesis of rRNA in the yeast Saccharomyces cerevisiae. Cold Spring Harb. Symp. Quant. Biol. 66, 555–565 (2001). - PubMed
-
- MacAlpine D. M., Bell S. P., A genomic view of eukaryotic DNA replication. Chromosome Res. 13, 309–326 (2005). - PubMed
-
- Fangman W. L., Brewer B. J., Activation of replication origins within yeast chromosomes. Annu. Rev. Cell Biol. 7, 375–402 (1991). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

