COPMAN: A novel high-throughput and highly sensitive method to detect viral nucleic acids including SARS-CoV-2 RNA in wastewater
- PMID: 36162583
- PMCID: PMC9502438
- DOI: 10.1016/j.scitotenv.2022.158966
COPMAN: A novel high-throughput and highly sensitive method to detect viral nucleic acids including SARS-CoV-2 RNA in wastewater
Abstract
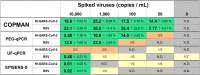
During the coronavirus disease 2019 (COVID-19) pandemic, wastewater-based epidemiology (WBE) attracted attention as an objective and comprehensive indicator of community infection that does not require individual inspection. Although several severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection methods from wastewater have been developed, there are obstacles to their social implementation. In this study, we developed the COPMAN (Coagulation and Proteolysis method using Magnetic beads for detection of Nucleic acids in wastewater), an automatable method that can concentrate and detect multiple types of viruses from a limited volume (∼10 mL) of wastewater. The COPMAN consists of a high basicity polyaluminum chloride (PAC) coagulation process, magnetic bead-based RNA purification, and RT-preamplification, followed by qPCR. A series of enzymes exhibiting a high tolerance to PCR inhibitors derived from wastewater was identified and employed in the molecular detection steps in the COPMAN. We compared the detectability of viral RNA from 10-mL samples of virus-spiked (heat-inactivated SARS-CoV-2 and intact RSV) or unspiked wastewater by the COPMAN and other methods (PEG-qPCR, UF-qPCR, and EPISENS-S). The COPMAN was the most efficient for detecting spiked viruses from wastewater, detecting the highest level of pepper mild mottle virus (PMMoV), a typical intrinsic virus in human stool, from wastewater samples. The COPMAN also successfully detected indigenous SARS-CoV-2 RNA from 12 samples of wastewater at concentrations of 2.2 × 104 to 5.4 × 105 copies/L, during initial stages of an infection wave in the right and the left bank of the Sagami River in Japan (0.65 to 11.45 daily reported cases per 100,000 people). These results indicate that the COPMAN is suitable for detection of multiple pathogens from small volume of wastewater in automated stations.
Keywords: Automation; COVID-19; Environmental surveillance; PCR; Sewage; Wastewater-based epidemiology.
Copyright © 2022 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Masaaki Kitajima reports financial support was provided by Shionogi & Co Ltd. Yuka Adachi Katayama reports a relationship with Shionogi & Co Ltd. that includes: employment. Shin Hayase reports a relationship with Shionogi & Co Ltd. that includes: employment. Yoshinori Ando reports a relationship with Shionogi & Co Ltd. that includes: employment. Tomohiro Kuroita reports a relationship with Shionogi & Co Ltd. that includes: employment. Kazuya Okada reports a relationship with Shionogi & Co Ltd. that includes: employment. Ryo Iwamoto reports a relationship with Shionogi & Co Ltd. that includes: employment. Toru Yanagimoto reports a relationship with Shionogi & Co Ltd. that includes: employment. Yusaku Masago reports a relationship with Shionogi & Co Ltd. that includes: employment. Yuka Adachi Katayama (Yuka Katayama) has patent Nucleic acid detection & quantification methods from environmental samples pending to Shionogi & Co Ltd., Hokkaido university. Shin Hayase has patent Nucleic acid detection & quantification methods from environmental samples pending to Shionogi & Co Ltd., Hokkaido university. Ryo Iwamoto has patent Nucleic acid detection and quantification methods from environmental samples pending to Shionogi & Co Ltd., Hokkaido University. Masaaki Kitajima has patent Nucleic acid detection and quantification methods from environmental samples pending to Shionogi & Co Ltd., Hokkaido University. Yusaku Masago has patent Nucleic acid detection and quantification methods from environmental samples pending to Shionogi & Co Ltd., Hokkaido university. Judy Noguchi had proofreaded this manuscript in English for a fee. Judy Noguchi works in Kobe Gakuin University.
Figures




Similar articles
-
Near full-automation of COPMAN using a LabDroid enables high-throughput and sensitive detection of SARS-CoV-2 RNA in wastewater as a leading indicator.Sci Total Environ. 2023 Jul 10;881:163454. doi: 10.1016/j.scitotenv.2023.163454. Epub 2023 Apr 13. Sci Total Environ. 2023. PMID: 37061063 Free PMC article.
-
Simultaneous extraction and detection of DNA and RNA from viruses, prokaryotes, and eukaryotes in wastewater using a modified COPMAN.Sci Total Environ. 2024 Jan 10;907:167866. doi: 10.1016/j.scitotenv.2023.167866. Epub 2023 Oct 19. Sci Total Environ. 2024. PMID: 37863234
-
The Efficient and Practical virus Identification System with ENhanced Sensitivity for Solids (EPISENS-S): A rapid and cost-effective SARS-CoV-2 RNA detection method for routine wastewater surveillance.Sci Total Environ. 2022 Oct 15;843:157101. doi: 10.1016/j.scitotenv.2022.157101. Epub 2022 Aug 8. Sci Total Environ. 2022. PMID: 35952875 Free PMC article.
-
SARS-CoV-2 detection and inactivation in water and wastewater: review on analytical methods, limitations and future research recommendations.Emerg Microbes Infect. 2023 Dec;12(2):2222850. doi: 10.1080/22221751.2023.2222850. Emerg Microbes Infect. 2023. PMID: 37279167 Free PMC article. Review.
-
Wastewater Based Epidemiology Perspective as a Faster Protocol for Detecting Coronavirus RNA in Human Populations: A Review with Specific Reference to SARS-CoV-2 Virus.Pathogens. 2021 Aug 10;10(8):1008. doi: 10.3390/pathogens10081008. Pathogens. 2021. PMID: 34451472 Free PMC article. Review.
Cited by
-
Genotype Diversity of Enteric Viruses in Wastewater Amid the COVID-19 Pandemic.Food Environ Virol. 2023 Jun;15(2):176-191. doi: 10.1007/s12560-023-09553-4. Epub 2023 Apr 14. Food Environ Virol. 2023. PMID: 37058225 Free PMC article.
-
Dissemination of local sub-variants of SARS-CoV-2 detected by detailed mutation analysis in wastewater-based epidemiology.PLoS One. 2025 May 28;20(5):e0317076. doi: 10.1371/journal.pone.0317076. eCollection 2025. PLoS One. 2025. PMID: 40434998 Free PMC article.
-
Wastewater-based reproduction numbers and projections of COVID-19 cases in three areas in Japan, November 2021 to December 2022.Euro Surveill. 2024 Feb;29(8):2300277. doi: 10.2807/1560-7917.ES.2024.29.8.2300277. Euro Surveill. 2024. PMID: 38390648 Free PMC article.
-
The Emergence and Widespread Circulation of Enteric Viruses Throughout the COVID-19 Pandemic: A Wastewater-Based Evidence.Food Environ Virol. 2023 Dec;15(4):342-354. doi: 10.1007/s12560-023-09566-z. Epub 2023 Oct 29. Food Environ Virol. 2023. PMID: 37898959
-
Protocol for detection of pathogenic enteric RNA viruses by regular monitoring of environmental samples from wastewater treatment plants using droplet digital PCR.Sci One Health. 2024 Oct 11;3:100080. doi: 10.1016/j.soh.2024.100080. eCollection 2024. Sci One Health. 2024. PMID: 39525942 Free PMC article.
References
-
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. - DOI - PMC - PubMed
-
- Ando H., Iwamoto R., Kobayashi H., Okabe S., Kitajima M. The efficient and practical virus identification system with ENhanced sensitivity for solids (EPISENS-S): a rapid and cost-effective SARS-CoV-2 RNA detection method for routine wastewater surveillance. Sci. Total Environ. 2022;843 doi: 10.1016/j.scitotenv.2022.157101. - DOI - PMC - PubMed
-
- Brouwer A.F., Eisenberg J.N.S., Pomeroy C.D., Shulman L.M., Hindiyeh M., Manor Y., Grotto I., Koopman J.S., Eisenberg M.C. Epidemiology of the silent polio outbreak in Rahat, Israel, based on modeling of environmental surveillance data. Proc. Natl. Acad. Sci. 2018;115 doi: 10.1073/pnas.1808798115. - DOI - PMC - PubMed
-
- COVID-19 Taskforce, Japan Society on Water Environment Manual for detection of SARS-CoV-2 RNA in wastewater. 2022. https://www.jswe.or.jp/aboutus/pdf/Manual%20for%20Detection%20of%20SARS-...
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

