A wheat resistosome defines common principles of immune receptor channels
- PMID: 36163289
- PMCID: PMC9581773
- DOI: 10.1038/s41586-022-05231-w
A wheat resistosome defines common principles of immune receptor channels
Abstract
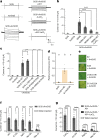
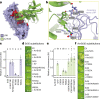
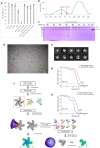
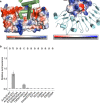
Plant intracellular nucleotide-binding leucine-rich repeat receptors (NLRs) detect pathogen effectors to trigger immune responses1. Indirect recognition of a pathogen effector by the dicotyledonous Arabidopsis thaliana coiled-coil domain containing NLR (CNL) ZAR1 induces the formation of a large hetero-oligomeric protein complex, termed the ZAR1 resistosome, which functions as a calcium channel required for ZAR1-mediated immunity2-4. Whether the resistosome and channel activities are conserved among plant CNLs remains unknown. Here we report the cryo-electron microscopy structure of the wheat CNL Sr355 in complex with the effector AvrSr356 of the wheat stem rust pathogen. Direct effector binding to the leucine-rich repeats of Sr35 results in the formation of a pentameric Sr35-AvrSr35 complex, which we term the Sr35 resistosome. Wheat Sr35 and Arabidopsis ZAR1 resistosomes bear striking structural similarities, including an arginine cluster in the leucine-rich repeats domain not previously recognized as conserved, which co-occurs and forms intramolecular interactions with the 'EDVID' motif in the coiled-coil domain. Electrophysiological measurements show that the Sr35 resistosome exhibits non-selective cation channel activity. These structural insights allowed us to generate new variants of closely related wheat and barley orphan NLRs that recognize AvrSr35. Our data support the evolutionary conservation of CNL resistosomes in plants and demonstrate proof of principle for structure-based engineering of NLRs for crop improvement.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures











Comment in
-
Wheeling in a new era in plant immunity.Nat Plants. 2022 Oct;8(10):1142-1143. doi: 10.1038/s41477-022-01257-0. Nat Plants. 2022. PMID: 36241732 No abstract available.
References
-
- Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

