Short prokaryotic Argonaute system repurposed as a nucleic acid detection tool
- PMID: 36163630
- PMCID: PMC9513045
- DOI: 10.1002/ctm2.1059
Short prokaryotic Argonaute system repurposed as a nucleic acid detection tool
Conflict of interest statement
Ana Potocnik and Daan C. Swarts (together with Balwina Koopal) submitted a patent application regarding the utilization of short pAgo systems for nucleic acid detection.
Figures
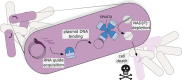

Comment on
-
Short prokaryotic Argonaute systems trigger cell death upon detection of invading DNA.Cell. 2022 Apr 28;185(9):1471-1486.e19. doi: 10.1016/j.cell.2022.03.012. Epub 2022 Apr 4. Cell. 2022. PMID: 35381200 Free PMC article.
Similar articles
-
Prokaryotic Argonaute Proteins: A New Frontier in Point-of-Care Viral Diagnostics.Int J Mol Sci. 2023 Oct 8;24(19):14987. doi: 10.3390/ijms241914987. Int J Mol Sci. 2023. PMID: 37834437 Free PMC article. Review.
-
Nucleic Acid-Binding Assay of Argonaute Protein Using Fluorescence Polarization.Methods Mol Biol. 2018;1680:123-129. doi: 10.1007/978-1-4939-7339-2_8. Methods Mol Biol. 2018. PMID: 29030845
-
Short prokaryotic Argonaute systems trigger cell death upon detection of invading DNA.Cell. 2022 Apr 28;185(9):1471-1486.e19. doi: 10.1016/j.cell.2022.03.012. Epub 2022 Apr 4. Cell. 2022. PMID: 35381200 Free PMC article.
-
Prokaryotic Argonaute proteins: novel genome-editing tools?Nat Rev Microbiol. 2018 Jan;16(1):5-11. doi: 10.1038/nrmicro.2017.73. Epub 2017 Jul 24. Nat Rev Microbiol. 2018. PMID: 28736447 Review.
-
Argonaute Proteins and Mechanisms of RNA Interference in Eukaryotes and Prokaryotes.Biochemistry (Mosc). 2018 May;83(5):483-497. doi: 10.1134/S0006297918050024. Biochemistry (Mosc). 2018. PMID: 29738683 Review.
Cited by
-
The nuclease-associated short prokaryotic Argonaute system nonspecifically degrades DNA upon activation by target recognition.Nucleic Acids Res. 2024 Jan 25;52(2):844-855. doi: 10.1093/nar/gkad1145. Nucleic Acids Res. 2024. PMID: 38048327 Free PMC article.
-
Prokaryotic Argonaute Proteins: A New Frontier in Point-of-Care Viral Diagnostics.Int J Mol Sci. 2023 Oct 8;24(19):14987. doi: 10.3390/ijms241914987. Int J Mol Sci. 2023. PMID: 37834437 Free PMC article. Review.
-
Oligomerization-mediated activation of a short prokaryotic Argonaute.Nature. 2023 Sep;621(7977):154-161. doi: 10.1038/s41586-023-06456-z. Epub 2023 Jul 26. Nature. 2023. PMID: 37494956 Free PMC article.
-
DNA-targeting short Argonautes complex with effector proteins for collateral nuclease activity and bacterial population immunity.Nat Microbiol. 2024 May;9(5):1368-1381. doi: 10.1038/s41564-024-01654-5. Epub 2024 Apr 15. Nat Microbiol. 2024. PMID: 38622379
References
-
- Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb Symp Quant Biol. 1986;51:263‐273. - PubMed
-
- Oliveira BB, Veigas B, Baptista PV. Isothermal Amplification of nucleic acids: the race for the next “Gold Standard”. Front Sens. 2011;2:752600.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
