A turn in species conservation for hairpin banksias: demonstration of oversplitting leads to better management of diversity
- PMID: 36164832
- PMCID: PMC9828017
- DOI: 10.1002/ajb2.16074
A turn in species conservation for hairpin banksias: demonstration of oversplitting leads to better management of diversity
Abstract
Premise: Understanding evolutionary history and classifying discrete units of organisms remain overwhelming tasks, and lags in this workload concomitantly impede an accurate documentation of biodiversity and conservation management. Rapid advances and improved accessibility of sensitive high-throughput sequencing tools are fortunately quickening the resolution of morphological complexes and thereby improving the estimation of species diversity. The recently described and critically endangered Banksia vincentia is morphologically similar to the hairpin banksia complex (B. spinulosa s.l.), a group of eastern Australian flowering shrubs whose continuum of morphological diversity has been responsible for taxonomic controversy and possibly questionable conservation initiatives.
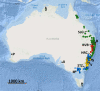
Methods: To assist conservation while testing the current taxonomy of this group, we used high-throughput sequencing to infer a population-scale evolutionary scenario for a sample set that is comprehensive in its representation of morphological diversity and a 2500-km distribution.
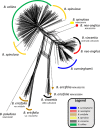
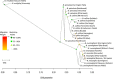
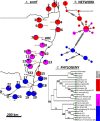
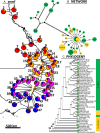
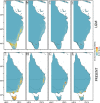
Results: Banksia spinulosa s.l. represents two clades, each with an internal genetic structure shaped through historical separation by biogeographic barriers. This structure conflicts with the existing taxonomy for the group. Corroboration between phylogeny and population statistics aligns with the hypothesis that B. collina, B. neoanglica, and B. vincentia should not be classified as species.
Conclusions: The pattern here supports how morphological diversity can be indicative of a locally expressed suite of traits rather than relationship. Oversplitting in the hairpin banksias is atypical since genomic analyses often reveal that species diversity is underestimated. However, we show that erring on overestimation can yield negative consequences, such as the disproportionate prioritization of a geographically anomalous population.
Keywords: Banksia; DArTseq; Proteaceae; cpDNA; genome-wide analysis; last glacial maximum; single nucleotide polymorphism; species boundaries; taxonomy; threatened species.
© 2022 State of New South Wales. American Journal of Botany published by Wiley Periodicals LLC on behalf of Botanical Society of America.
Conflict of interest statement
All authors declare no conflict of interest.
Figures

References
-
- Abbott, I. 1985. Reproductive ecology of Banksia grandis (Proteaceae). New Phytologist 99: 129–148.
-
- Acosta, M. C. , and Premoli A. C.. 2010. Evidence of chloroplast capture in South American Nothofagus (subgenus Nothofagus, Nothofagaceae). Molecular Phylogenetics and Evolution 54: 235–242. - PubMed
-
- Ahrens, C. W. , James E. A., Miller A. D., Scott F., Aitken N. C., Jones A. W., Lu‐Irving P., et al. 2020a. Spatial, climate and ploidy factors drive genomic diversity and resilience in the widespread grass Themeda triandra . Molecular Ecology 29: 3872–3888. - PubMed
-
- Australian National Botanical Garden . 2022. Australian Plant Name Index (APNI), Vascular plants, Banksia spinulosa [online; part of IBIS database]. Website: https://biodiversity.org.au/nsl/services/search/names?product=APNIandtre... [accessed 22 February 2022]. Australian National Botanical Garden and Centre for Australian National Biodiversity Research, Canberra, Australia.
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources

