Transcriptomic Profiling Identifies CD8+ T Cells in the Brain of Aged and Alzheimer's Disease Transgenic Mice as Tissue-Resident Memory T Cells
- PMID: 36165202
- PMCID: PMC9515311
- DOI: 10.4049/jimmunol.2100737
Transcriptomic Profiling Identifies CD8+ T Cells in the Brain of Aged and Alzheimer's Disease Transgenic Mice as Tissue-Resident Memory T Cells
Abstract
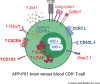
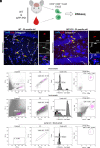
Peripheral immune cell infiltration into the brain is a prominent feature in aging and various neurodegenerative diseases such as Alzheimer's disease (AD). As AD progresses, CD8+ T cells infiltrate into the brain parenchyma, where they tightly associate with neurons and microglia. The functional properties of CD8+ T cells in the brain are largely unknown. To gain further insights into the putative functions of CD8+ T cells in the brain, we explored and compared the transcriptomic profile of CD8+ T cells isolated from the brain and blood of transgenic AD (APPswe/PSEN1dE9, line 85 [APP-PS1]) and age-matched wild-type (WT) mice. Brain CD8+ T cells of APP-PS1 and WT animals had similar transcriptomic profiles and substantially differed from blood circulating CD8+ T cells. The gene signature of brain CD8+ T cells identified them as tissue-resident memory (Trm) T cells. Gene Ontology enrichment and Kyoto Encyclopedia of Genes and Genomes pathway analysis on the significantly upregulated genes revealed overrepresentation of biological processes involved in IFN-β signaling and the response to viral infections. Furthermore, brain CD8+ T cells of APP-PS1 and aged WT mice showed similar differentially regulated genes as brain Trm CD8+ T cells in mouse models with acute virus infection, chronic parasite infection, and tumor growth. In conclusion, our profiling of brain CD8+ T cells suggests that in AD, these cells exhibit similar adaptive immune responses as in other inflammatory diseases of the CNS, potentially opening the door for immunotherapy in AD.
Copyright © 2022 by The American Association of Immunologists, Inc.
Conflict of interest statement
The authors have no financial conflicts of interest.
Figures





References
-
- Querfurth H. W., LaFerla F. M.. 2010. Alzheimer’s disease. N. Engl. J. Med. 362: 329–344. - PubMed
-
- Goedert M., Spillantini M. G.. 2006. A century of Alzheimer’s disease. Science 314: 777–781. - PubMed
-
- Brookmeyer R., Johnson E., Ziegler-Graham K., Arrighi H. M.. 2007. Forecasting the global burden of Alzheimer’s disease. Alzheimers Dement. 3: 186–191. - PubMed
-
- Alzheimer’s Association . 2015. 2015 Alzheimer’s disease facts and figures. Alzheimers Dement. 11: 332–384. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

