Hybrid Vigor: Importance of Hybrid λ Phages in Early Insights in Molecular Biology
- PMID: 36165780
- PMCID: PMC9799177
- DOI: 10.1128/mmbr.00124-21
Hybrid Vigor: Importance of Hybrid λ Phages in Early Insights in Molecular Biology
Abstract
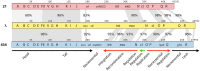
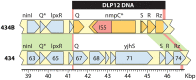
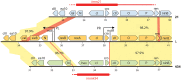
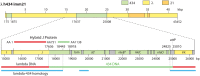
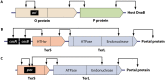
Laboratory-generated hybrids between phage λ and related phages played a seminal role in establishment of the λ model system, which, in turn, served to develop many of the foundational concepts of molecular biology, including gene structure and control. Important λ hybrids with phages 21 and 434 were the earliest of such phages. To understand the biology of these hybrids in full detail, we determined the complete genome sequences of phages 21 and 434. Although both genomes are canonical members of the λ-like phage family, they both carry unsuspected bacterial virulence gene types not previously described in this group of phages. In addition, we determined the sequences of the hybrid phages λ imm21, λ imm434, and λ h434 imm21. These sequences show that the replacements of λ DNA by nonhomologous segments of 21 or 434 DNA occurred through homologous recombination in adjacent sequences that are nearly identical in the parental phages. These five genome sequences correct a number of errors in published sequence fragments of the 21 and 434 genomes, and they point out nine nucleotide differences from Sanger's original λ sequence that are likely present in most extant λ strains in laboratory use today. We discuss the historical importance of these hybrid phages in the development of fundamental tenets of molecular biology and in some of the earliest gene cloning vectors. The 434 and 21 genomes reinforce the conclusion that the genomes of essentially all natural λ-like phages are mosaics of sequence modules from a pool of exchangeable segments.
Keywords: bacteriophage; hybrid phage; lysogeny; phage; phage 21; phage 434; phage genetics; phage lambda; temperate phage.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Aberrant immunity behaviour of hybrid lambda imm21 phages containing the DNA of ColE1-type plasmids.Mol Gen Genet. 1979;172(3):329-37. doi: 10.1007/BF00271733. Mol Gen Genet. 1979. PMID: 45615
-
Temperate phages acquire DNA from defective prophages by relaxed homologous recombination: the role of Rad52-like recombinases.PLoS Genet. 2014 Mar 6;10(3):e1004181. doi: 10.1371/journal.pgen.1004181. eCollection 2014 Mar. PLoS Genet. 2014. PMID: 24603854 Free PMC article.
-
Population Dynamics of Phage and Bacteria in Spatially Structured Habitats Using Phage λ and Escherichia coli.J Bacteriol. 2016 May 27;198(12):1783-93. doi: 10.1128/JB.00965-15. Print 2016 Jun 15. J Bacteriol. 2016. PMID: 27068593 Free PMC article.
-
Comparative molecular biology of lambdoid phages.Annu Rev Microbiol. 1994;48:193-222. doi: 10.1146/annurev.mi.48.100194.001205. Annu Rev Microbiol. 1994. PMID: 7826005 Review.
-
Bacteriophage lambda: Early pioneer and still relevant.Virology. 2015 May;479-480:310-30. doi: 10.1016/j.virol.2015.02.010. Epub 2015 Mar 3. Virology. 2015. PMID: 25742714 Free PMC article. Review.
Cited by
-
Lambdoid phages with abundant Chi recombination hotspots reflect diverse viral strategies for recombination-dependent growth.Genome Res. 2025 Aug 1;35(8):1767-1780. doi: 10.1101/gr.280248.124. Genome Res. 2025. PMID: 40639916 Free PMC article.
-
Genomic analysis of Anderson typing phages of Salmonella Typhimrium: towards understanding the basis of bacteria-phage interaction.Sci Rep. 2023 Jun 28;13(1):10484. doi: 10.1038/s41598-023-37307-6. Sci Rep. 2023. PMID: 37380724 Free PMC article.
-
Research on phage λ: a lucky choice.EcoSal Plus. 2024 Dec 12;12(1):eesp00142023. doi: 10.1128/ecosalplus.esp-0014-2023. Epub 2023 Dec 22. EcoSal Plus. 2024. PMID: 39665539 Free PMC article. Review.
References
-
- Lederberg EM. 1950. Lysogenicity in Escherichia coli strain K-12. Microb Genet Bull 1:5–9.
-
- Jacob F, Wollman EL. 1961. Sexuality and the genetics of bacteria. Academic Press, New York, NY.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

