Stromal Reprogramming by FAK Inhibition Overcomes Radiation Resistance to Allow for Immune Priming and Response to Checkpoint Blockade
- PMID: 36165893
- PMCID: PMC9722639
- DOI: 10.1158/2159-8290.CD-22-0192
Stromal Reprogramming by FAK Inhibition Overcomes Radiation Resistance to Allow for Immune Priming and Response to Checkpoint Blockade
Abstract
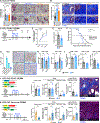
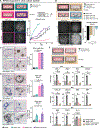
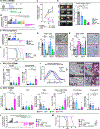
The effects of radiotherapy (RT) on tumor immunity in pancreatic ductal adenocarcinoma (PDAC) are not well understood. To better understand if RT can prime antigen-specific T-cell responses, we analyzed human PDAC tissues and mouse models. In both settings, there was little evidence of RT-induced T-cell priming. Using in vitro systems, we found that tumor-stromal components, including fibroblasts and collagen, cooperate to blunt RT efficacy and impair RT-induced interferon signaling. Focal adhesion kinase (FAK) inhibition rescued RT efficacy in vitro and in vivo, leading to tumor regression, T-cell priming, and enhanced long-term survival in PDAC mouse models. Based on these data, we initiated a clinical trial of defactinib in combination with stereotactic body RT in patients with PDAC (NCT04331041). Analysis of PDAC tissues from these patients showed stromal reprogramming mirroring our findings in genetically engineered mouse models. Finally, the addition of checkpoint immunotherapy to RT and FAK inhibition in animal models led to complete tumor regression and long-term survival.
Significance: Checkpoint immunotherapeutics have not been effective in PDAC, even when combined with RT. One possible explanation is that RT fails to prime T-cell responses in PDAC. Here, we show that FAK inhibition allows RT to prime tumor immunity and unlock responsiveness to checkpoint immunotherapy. This article is highlighted in the In This Issue feature, p. 2711.
©2022 American Association for Cancer Research.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- F30 CA254087/CA/NCI NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- R01 CA203890/CA/NCI NIH HHS/United States
- R01 CA244938/CA/NCI NIH HHS/United States
- R01 CA248917/CA/NCI NIH HHS/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- P50 CA196510/CA/NCI NIH HHS/United States
- P30 DK052574/DK/NIDDK NIH HHS/United States
- UL1 TR002345/TR/NCATS NIH HHS/United States
- R50 CA221675/CA/NCI NIH HHS/United States
- T32 EB021955/EB/NIBIB NIH HHS/United States
- F31 DK122633/DK/NIDDK NIH HHS/United States
- T32 GM007200/GM/NIGMS NIH HHS/United States
- R01 CA262506/CA/NCI NIH HHS/United States
- F30 CA243233/CA/NCI NIH HHS/United States
- R01 CA177670/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

