Exploratory genomic analysis of high-grade neuroendocrine neoplasms across diverse primary sites
- PMID: 36165930
- PMCID: PMC10043760
- DOI: 10.1530/ERC-22-0015
Exploratory genomic analysis of high-grade neuroendocrine neoplasms across diverse primary sites
Abstract
High-grade (grade 3) neuroendocrine neoplasms (G3 NENs) have poor survival outcomes. From a clinical standpoint, G3 NENs are usually grouped regardless of primary site and treated similarly. Little is known regarding the underlying genomics of these rare tumors, especially when compared across different primary sites. We performed whole transcriptome (n = 46), whole exome (n = 40), and gene copy number (n = 43) sequencing on G3 NEN formalin-fixed, paraffin-embedded samples from diverse organs (in total, 17 were lung, 16 were gastroenteropancreatic, and 13 other). G3 NENs despite arising from diverse primary sites did not have gene expression profiles that were easily segregated by organ of origin. Across all G3 NENs, TP53, APC, RB1, and CDKN2A were significantly mutated. The CDK4/6 cell cycling pathway was mutated in 95% of cases, with upregulation of oncogenes within this pathway. G3 NENs had high tumor mutation burden (mean 7.09 mutations/MB), with 20% having >10 mutations/MB. Two somatic copy number alterations were significantly associated with worse prognosis across tissue types: focal deletion 22q13.31 (HR, 7.82; P = 0.034) and arm amplification 19q (HR, 4.82; P = 0.032). This study is among the most diverse genomic study of high-grade neuroendocrine neoplasms. We uncovered genomic features previously unrecognized for this rapidly fatal and rare cancer type that could have potential prognostic and therapeutic implications.
Keywords: APC; CDK4; CDKN2A; RB1; TP53; cell cycling; genomics; grade 3; high grade; neuroendocrine carcinoma; neuroendocrine neoplasm; neuroendocrine tumor; sequencing; tumor mutation burden.
Conflict of interest statement
Conflicts of Interest
The remaining authors declare no conflicts of interest.
Figures



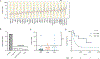

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

