Machine Learning in Nutrition Research
- PMID: 36166846
- PMCID: PMC9776646
- DOI: 10.1093/advances/nmac103
Machine Learning in Nutrition Research
Erratum in
-
Corrigendum to "Machine Learning in Nutrition Research".Adv Nutr. 2023 May;14(3):584. doi: 10.1016/j.advnut.2023.03.001. Epub 2023 Mar 10. Adv Nutr. 2023. PMID: 36913977 Free PMC article. No abstract available.
-
Corrigendum to "Machine Learning in Nutrition Research".Adv Nutr. 2023 Apr 1:S2161-8313(23)00281-8. doi: 10.1016/j.advnut.2023.03.012. Online ahead of print. Adv Nutr. 2023. PMID: 37011883 No abstract available.
Abstract
Data currently generated in the field of nutrition are becoming increasingly complex and high-dimensional, bringing with them new methods of data analysis. The characteristics of machine learning (ML) make it suitable for such analysis and thus lend itself as an alternative tool to deal with data of this nature. ML has already been applied in important problem areas in nutrition, such as obesity, metabolic health, and malnutrition. Despite this, experts in nutrition are often without an understanding of ML, which limits its application and therefore potential to solve currently open questions. The current article aims to bridge this knowledge gap by supplying nutrition researchers with a resource to facilitate the use of ML in their research. ML is first explained and distinguished from existing solutions, with key examples of applications in the nutrition literature provided. Two case studies of domains in which ML is particularly applicable, precision nutrition and metabolomics, are then presented. Finally, a framework is outlined to guide interested researchers in integrating ML into their work. By acting as a resource to which researchers can refer, we hope to support the integration of ML in the field of nutrition to facilitate modern research.
Keywords: XGBoost; cardiovascular disease; diabetes; machine learning; models; obesity; omics; personalized nutrition; random forest.
© The Author(s) 2022. Published by Oxford University Press on behalf of the American Society for Nutrition.
Figures
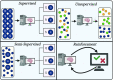

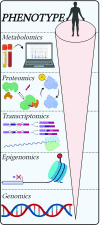
References
-
- Reel PS, Reel S, Pearson E, Trucco E, Jefferson E. Using machine learning approaches for multi-omics data analysis: a review. Biotechnol Adv. 2021;49:107739. - PubMed
-
- Li R, Li L, Xu Y, Yang J. Machine learning meets omics: applications and perspectives. Brief Bioinform. 2022;23(1):bbab460. - PubMed
-
- Wang X, Liu J, Ma L. Identification of gut flora based on robust support vector machine. J Phys Conf Ser. 2022;2171(1):012066.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

