Carboxypeptidase G and pterin deaminase metabolic pathways degrade folic acid in Variovorax sp. F1
- PMID: 36167524
- PMCID: PMC9513972
- DOI: 10.1186/s12866-022-02643-6
Carboxypeptidase G and pterin deaminase metabolic pathways degrade folic acid in Variovorax sp. F1
Abstract
Background: Folic acid (FA) is a synthetic vitamin (B9) and the oxidized form of a metabolic cofactor that is essential for life. Although the biosynthetic mechanisms of FA are established, its environmental degradation mechanism has not been fully elucidated. The present study aimed to identify bacteria in soil that degrade FA and the mechanisms involved.
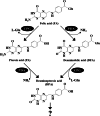
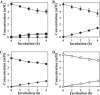
Results: We isolated the soil bacterium Variovorax sp. F1 from sampled weed rhizospheres in a grassland and investigated its FA degradation mechanism. Cultured Variovorax sp. F1 rapidly degraded FA to pteroic acid (PA), indicating that FA hydrolysis to PA and glutamate. We cloned the carboxypeptidase G (CPG) gene and found widely distributed paralogs within the Variovorax genus. Recombinant CPG preferred FA and deaminofolic acid as substrates, indicating its involvement in FA degradation by Variovorax. Prolonged culture of Variovorax sp. F1 resulted in decreased rates of deaminofolic acid (DFA) and deaminopteroic acid (DPA) accumulation. This indicated that the deamination reaction also comprised a route of FA degradation. We also identified an F1 gene that was orthologous to the pterin deaminase gene (Arad3529) of Agrobacterium radiobacter. The encoded protein deaminated FA and PA to DFA and DPA, which was consistent with the deamination activity of FA and PA in bacterial cell-free extracts.
Conclusion: We discovered that the two enzymes required for FA degradation pathways in isolates of Variovorax sp. F1 comprise CPG and pterin deaminase, and that DFA and PA are intermediates in the generation of DPA.
Keywords: Deaminofolic acid; Pterin deaminase; Pteroic acid; Variovorax; Vitamin B9.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- McDowell LR. Vitamins in animal and human nutrition. 2. Ames: Iowa State University Press; 2000.
-
- Shenkin A. Basics in clinical nutrition: physiological function and deficiency states of vitamins. Eur J Clin Nutr Metab. 2008;3:e275–e280. doi: 10.1016/j.eclnm.2008.06.008. - DOI
-
- Parker GL, Smith LK, Baxendale IR. Development of the industrial synthesis of vitamin a. Tetrahedron. 2016;72:1645–1652. doi: 10.1016/j.tet.2016.02.029. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

