Metabolic control of histone acetylation for precise and timely regulation of minor ZGA in early mammalian embryos
- PMID: 36167681
- PMCID: PMC9515074
- DOI: 10.1038/s41421-022-00440-z
Metabolic control of histone acetylation for precise and timely regulation of minor ZGA in early mammalian embryos
Abstract
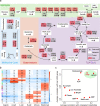
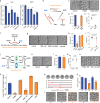
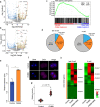
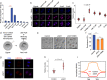
Metabolism feeds into the regulation of epigenetics via metabolic enzymes and metabolites. However, metabolic features, and their impact on epigenetic remodeling during mammalian pre-implantation development, remain poorly understood. In this study, we established the metabolic landscape of mouse pre-implantation embryos from zygote to blastocyst, and quantified some absolute carbohydrate metabolites. We integrated these data with transcriptomic and proteomic data, and discovered the metabolic characteristics of the development process, including the activation of methionine cycle from 8-cell embryo to blastocyst, high glutaminolysis metabolism at blastocyst stage, enhanced TCA cycle activity from the 8-cell embryo stage, and active glycolysis in the blastocyst. We further demonstrated that oxidized nicotinamide adenine dinucleotide (NAD+) synthesis is indispensable for mouse pre-implantation development. Mechanistically, in part, NAD+ is required for the exit of minor zygotic gene activation (ZGA) by cooperating with SIRT1 to remove zygotic H3K27ac. In human, NAD+ supplement can promote the removal of zygotic H3K27ac and benefit pre-implantation development. Our findings demonstrate that precise and timely regulation of minor ZGA is controlled by metabolic dynamics, and enhance our understanding of the metabolism of mammalian early embryos.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

