The ecology and epidemiology of malaria parasitism in wild chimpanzee reservoirs
- PMID: 36167977
- PMCID: PMC9515101
- DOI: 10.1038/s42003-022-03962-0
The ecology and epidemiology of malaria parasitism in wild chimpanzee reservoirs
Abstract
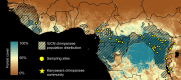
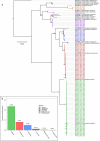
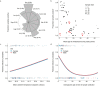
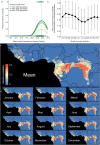
Chimpanzees (Pan troglodytes) harbor rich assemblages of malaria parasites, including three species closely related to P. falciparum (sub-genus Laverania), the most malignant human malaria parasite. Here, we characterize the ecology and epidemiology of malaria infection in wild chimpanzee reservoirs. We used molecular assays to screen chimpanzee fecal samples, collected longitudinally and cross-sectionally from wild populations, for malaria parasite mitochondrial DNA. We found that chimpanzee malaria parasitism has an early age of onset and varies seasonally in prevalence. A subset of samples revealed Hepatocystis mitochondrial DNA, with phylogenetic analyses suggesting that Hepatocystis appears to cross species barriers more easily than Laverania. Longitudinal and cross-sectional sampling independently support the hypothesis that mean ambient temperature drives spatiotemporal variation in chimpanzee Laverania infection. Infection probability peaked at ~24.5 °C, consistent with the empirical transmission optimum of P. falciparum in humans. Forest cover was also positively correlated with spatial variation in Laverania prevalence, consistent with the observation that forest-dwelling Anophelines are the primary vectors. Extrapolating these relationships across equatorial Africa, we map spatiotemporal variation in the suitability of chimpanzee habitat for Laverania transmission, offering a hypothetical baseline indicator of human exposure risk.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Liu, W. et al. Single genome amplification and direct amplicon sequencing of Plasmodium spp. DNA from ape fecal specimens. Protocol Exchange 1–14 (2010).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

