Reverse Transcription-Loop-Mediated Isothermal Amplification-CRISPR-Cas13a Technology as a Promising Diagnostic Tool for SARS-CoV-2
- PMID: 36169448
- PMCID: PMC9604158
- DOI: 10.1128/spectrum.02398-22
Reverse Transcription-Loop-Mediated Isothermal Amplification-CRISPR-Cas13a Technology as a Promising Diagnostic Tool for SARS-CoV-2
Abstract
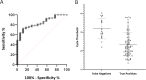
At the end of 2019, a new coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), caused a pandemic that persists to date and has resulted in more than 6.2 million deaths. In the last couple of years, researchers have made great efforts to develop a diagnostic technique that maintains high levels of sensitivity and specificity, since an accurate and early diagnosis is required to minimize the prevalence of SARS-CoV-2 infection. In this context, CRISPR-Cas systems are proposed as promising tools for development as diagnostic techniques due to their high specificity, highlighting that Cas13 endonuclease discriminates single nucleotide changes and displays collateral activity against single-stranded RNA molecules. With the aim of improving the sensitivity of diagnosis, this technology is usually combined with isothermal preamplification reactions (SHERLOCK, DETECTR). Based on this, we developed a reverse transcription-loop-mediated isothermal amplification (RT-LAMP)-CRISPR-Cas13a method for SARS-CoV-2 virus detection in nasopharyngeal samples without using RNA extraction that exhibits 100% specificity and 83% sensitivity, as well as a positive predictive value (PPV) of 100% and negative predictive values (NPVs) of 100%, 81%, 79.1%, and 66.7% for cycle threshold (CT) values of <20, 20 to 30, >30 and overall, respectively. IMPORTANCE The coronavirus disease 2019 (COVID-19) crisis has driven the development of innovative molecular diagnosis methods, including CRISPR-Cas technology. In this work, we performed a protocol, working with RNA extraction kit-free samples and using RT-LAMP-CRISPR-Cas13a technology; our results place this method at the forefront of rapid and specific diagnostic methods for COVID-19 due to the high specificity (100%), sensitivity (83%), PPVs (100%), and NPVs (81% for high viral loads) obtained with clinical samples.
Keywords: COVID-19; CRISPR-Cas; CRISPR-Cas13; RT-LAMP; SARS-CoV-2; diagnosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
CLEVER assay: A visual and rapid RNA extraction-free detection of SARS-CoV-2 based on CRISPR-Cas integrated RT-LAMP technology.J Appl Microbiol. 2022 Aug;133(2):410-421. doi: 10.1111/jam.15571. Epub 2022 Apr 18. J Appl Microbiol. 2022. PMID: 35396760 Free PMC article.
-
iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2.Virus Res. 2020 Oct 15;288:198129. doi: 10.1016/j.virusres.2020.198129. Epub 2020 Aug 18. Virus Res. 2020. PMID: 32822689 Free PMC article.
-
Development and Validation of a Novel COVID-19 nsp8 One-Tube RT-LAMP-CRISPR Assay for SARS-CoV-2 Diagnosis.Microbiol Spectr. 2022 Dec 21;10(6):e0196222. doi: 10.1128/spectrum.01962-22. Epub 2022 Nov 29. Microbiol Spectr. 2022. PMID: 36445095 Free PMC article.
-
Diagnostic efficiency of RT-LAMP integrated CRISPR-Cas technique for COVID-19: A systematic review and meta-analysis.Pathog Glob Health. 2022 Oct;116(7):410-420. doi: 10.1080/20477724.2022.2035625. Epub 2022 Feb 10. Pathog Glob Health. 2022. PMID: 35142264 Free PMC article.
-
A Recent Update on Advanced Molecular Diagnostic Techniques for COVID-19 Pandemic: An Overview.Front Immunol. 2021 Dec 14;12:732756. doi: 10.3389/fimmu.2021.732756. eCollection 2021. Front Immunol. 2021. PMID: 34970254 Free PMC article. Review.
Cited by
-
A Comprehensive Comparison of Rapid RNA Extraction Methods for Detection of SARS-CoV-2 as the Infectious Agent of the Upper Respiratory Tract using Direct RT-LAMP Assay.Adv Biomed Res. 2023 Nov 29;12:261. doi: 10.4103/abr.abr_63_23. eCollection 2023. Adv Biomed Res. 2023. PMID: 38192891 Free PMC article.
-
The LAMP-CRISPR-Cas13a technique for detecting the CBASS mechanism of phage resistance in bacteria.Front Microbiol. 2025 Mar 24;16:1550534. doi: 10.3389/fmicb.2025.1550534. eCollection 2025. Front Microbiol. 2025. PMID: 40196034 Free PMC article.
-
Immunological Studies to Understand Hybrid/Recombinant Variants of SARS-CoV-2.Vaccines (Basel). 2022 Dec 25;11(1):45. doi: 10.3390/vaccines11010045. Vaccines (Basel). 2022. PMID: 36679891 Free PMC article. Review.
-
Rapid molecular detection of Senecavirus A based on reverse transcription loop-mediated isothermal amplification (RT-LAMP) and CRISPR/Cas12a.Front Bioeng Biotechnol. 2025 Apr 4;13:1451125. doi: 10.3389/fbioe.2025.1451125. eCollection 2025. Front Bioeng Biotechnol. 2025. PMID: 40256781 Free PMC article.
-
Loop-Mediated Isothermal Amplification-Integrated CRISPR Methods for Infectious Disease Diagnosis at Point of Care.ACS Omega. 2023 Nov 7;8(46):43357-43373. doi: 10.1021/acsomega.3c04422. eCollection 2023 Nov 21. ACS Omega. 2023. PMID: 38027359 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

