GWAS in the southern African context
- PMID: 36170230
- PMCID: PMC9518849
- DOI: 10.1371/journal.pone.0264657
GWAS in the southern African context
Abstract
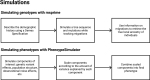
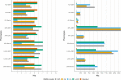
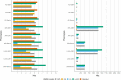
Researchers would generally adjust for the possible confounding effect of population structure by considering global ancestry proportions or top principle components. Alternatively, researchers would conduct admixture mapping to increase the power to detect variants with an ancestry effect. This is sufficient in simple admixture scenarios, however, populations from southern Africa can be complex multi-way admixed populations. Duan et al. (2018) first described local ancestry adjusted allelic (LAAA) analysis as a robust method for discovering association signals, while producing minimal false positive hits. Their simulation study, however, was limited to a two-way admixed population. Realizing that their findings might not translate to other admixture scenarios, we simulated a three- and five-way admixed population to compare the LAAA model to other models commonly used in genome-wide association studies (GWAS). We found that, given our admixture scenarios, the LAAA model identifies the most causal variants in most of the phenotypes we tested across both the three-way and five-way admixed populations. The LAAA model also produced a high number of false positive hits which was potentially caused by the ancestry effect size that we assumed. Considering the extent to which the various models tested differed in their results and considering that the source of a given association is unknown, we recommend that researchers use multiple GWAS models when analysing populations with complex ancestry.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

