Accurate detection of benign and malignant renal tumor subtypes with MethylBoostER: An epigenetic marker-driven learning framework
- PMID: 36170366
- PMCID: PMC9519038
- DOI: 10.1126/sciadv.abn9828
Accurate detection of benign and malignant renal tumor subtypes with MethylBoostER: An epigenetic marker-driven learning framework
Abstract
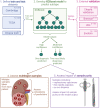
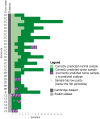
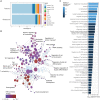
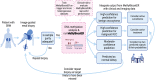
Current gold standard diagnostic strategies are unable to accurately differentiate malignant from benign small renal masses preoperatively; consequently, 20% of patients undergo unnecessary surgery. Devising a more confident presurgical diagnosis is key to improving treatment decision-making. We therefore developed MethylBoostER, a machine learning model leveraging DNA methylation data from 1228 tissue samples, to classify pathological subtypes of renal tumors (benign oncocytoma, clear cell, papillary, and chromophobe RCC) and normal kidney. The prediction accuracy in the testing set was 0.960, with class-wise ROC AUCs >0.988 for all classes. External validation was performed on >500 samples from four independent datasets, achieving AUCs >0.89 for all classes and average accuracies of 0.824, 0.703, 0.875, and 0.894 for the four datasets. Furthermore, consistent classification of multiregion samples (N = 185) from the same patient demonstrates that methylation heterogeneity does not limit model applicability. Following further clinical studies, MethylBoostER could facilitate a more confident presurgical diagnosis to guide treatment decision-making in the future.
Figures




References
-
- Shuch B., Amin A., Armstrong A. J., Eble J. N., Ficarra V., Lopez-Beltran A., Martignoni G., Rini B. I., Kutikov A., Understanding pathologic variants of renal cell carcinoma: Distilling therapeutic opportunities from biologic complexity. Eur. Urol. 67, 85–97 (2015). - PubMed
-
- Moch H., Cubilla A. L., Humphrey P. A., Reuter V. E., Ulbright T. M., The 2016 WHO classification of tumours of the urinary system and male genital organs-part A: Renal, penile, and testicular tumours. Eur. Urol. 70, 93–105 (2016). - PubMed
-
- Patel H. D., Druskin S. C., Rowe S. P., Pierorazio P. M., Gorin M. A., Allaf M. E., Surgical histopathology for suspected oncocytoma on renal mass biopsy: A systematic review and meta-analysis. BJU Int. 119, 661–666 (2017). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

