Circadian disruption enhances HSF1 signaling and tumorigenesis in Kras-driven lung cancer
- PMID: 36170373
- PMCID: PMC9519049
- DOI: 10.1126/sciadv.abo1123
Circadian disruption enhances HSF1 signaling and tumorigenesis in Kras-driven lung cancer
Abstract
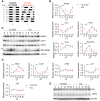
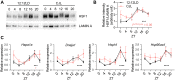
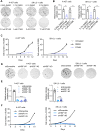
Disrupted circadian rhythmicity is a prominent feature of modern society and has been designated as a probable carcinogen by the World Health Organization. However, the biological mechanisms that connect circadian disruption and cancer risk remain largely undefined. We demonstrate that exposure to chronic circadian disruption [chronic jetlag (CJL)] increases tumor burden in a mouse model of KRAS-driven lung cancer. Molecular characterization of tumors and tumor-bearing lung tissues revealed that CJL enhances the expression of heat shock factor 1 (HSF1) target genes. Consistently, exposure to CJL disrupted the highly rhythmic nuclear trafficking of HSF1 in the lung, resulting in an enhanced accumulation of HSF1 in the nucleus. HSF1 has been shown to promote tumorigenesis in other systems, and we find that pharmacological or genetic inhibition of HSF1 reduces the growth of KRAS-mutant human lung cancer cells. These findings implicate HSF1 as a molecular link between circadian disruption and enhanced tumorigenesis.
Figures



References
-
- N. O. H. S. (NHIS-OHS, 2015); wwwn.cdc.gov/Niosh-whc/chart/ohs-workorg?OU=WORKSCHD_RCD&T=I&V=R2.
-
- Ye Y., Xiang Y., Ozguc F. M., Kim Y., Liu C.-J., Park P. K., Hu Q., Diao L., Lou Y., Lin C., Guo A.-Y., Zhou B., Wang L., Chen Z., Takahashi J. S., Mills G. B., Yoo S.-H., Han L., The genomic landscape and pharmacogenomic interactions of clock genes in cancer chronotherapy. Cell Syst 6, 314–328.e2 (2018). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

