Single-cell transcriptomics identifies pathogenic T-helper 17.1 cells and pro-inflammatory monocytes in immune checkpoint inhibitor-related pneumonitis
- PMID: 36171010
- PMCID: PMC9528720
- DOI: 10.1136/jitc-2022-005323
Single-cell transcriptomics identifies pathogenic T-helper 17.1 cells and pro-inflammatory monocytes in immune checkpoint inhibitor-related pneumonitis
Abstract
Background: Immune checkpoint inhibitor (ICI)-related pneumonitis is the most frequent fatal immune-related adverse event associated with programmed cell death protein-1/programmed death ligand-1 blockade. The pathophysiology however remains largely unknown, owing to limited and contradictory findings in existing literature pointing at either T-helper 1 or T-helper 17-mediated autoimmunity. In this study, we aimed to gain novel insights into the mechanisms of ICI-related pneumonitis, thereby identifying potential therapeutic targets.
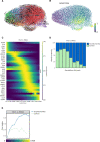
Methods: In this prospective observational study, single-cell RNA and T-cell receptor sequencing was performed on bronchoalveolar lavage fluid of 11 patients with ICI-related pneumonitis and 6 demographically-matched patients with cancer without ICI-related pneumonitis. Single-cell transcriptomic immunophenotyping and cell fate mapping coupled to T-cell receptor repertoire analyses were performed.
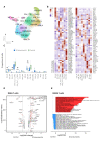
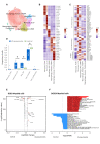
Results: We observed enrichment of both CD4+ and CD8+ T cells in ICI-pneumonitis bronchoalveolar lavage fluid. The CD4+ T-cell compartment showed an increase of pathogenic T-helper 17.1 cells, characterized by high co-expression of TBX21 (encoding T-bet) and RORC (ROR-γ), IFN-G (IFN-γ), IL-17A, CSF2 (GM-CSF), and cytotoxicity genes. Type 1 regulatory T cells and naïve-like CD4+ T cells were also enriched. Within the CD8+ T-cell compartment, mainly effector memory T cells were increased. Correspondingly, myeloid cells in ICI-pneumonitis bronchoalveolar lavage fluid were relatively depleted of anti-inflammatory resident alveolar macrophages while pro-inflammatory 'M1-like' monocytes (expressing TNF, IL-1B, IL-6, IL-23A, and GM-CSF receptor CSF2RA, CSF2RB) were enriched compared with control samples. Importantly, a feedforward loop, in which GM-CSF production by pathogenic T-helper 17.1 cells promotes tissue inflammation and IL-23 production by pro-inflammatory monocytes and vice versa, has been well characterized in multiple autoimmune disorders but has never been identified in ICI-related pneumonitis.
Conclusions: Using single-cell transcriptomics, we identified accumulation of pathogenic T-helper 17.1 cells in ICI-pneumonitis bronchoalveolar lavage fluid-a phenotype explaining previous divergent findings on T-helper 1 versus T-helper 17 involvement in ICI-pneumonitis-,putatively engaging in detrimental crosstalk with pro-inflammatory 'M1-like' monocytes. This finding yields several novel potential therapeutic targets for the treatment of ICI-pneumonitis. Most notably repurposing anti-IL-23 merits further research as a potential efficacious and safe treatment for ICI-pneumonitis.
Keywords: Autoimmunity; Computational Biology; Immunotherapy; Lung Neoplasms; Programmed Cell Death 1 Receptor.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
