Visualizing translation dynamics at atomic detail inside a bacterial cell
- PMID: 36171285
- PMCID: PMC9534751
- DOI: 10.1038/s41586-022-05255-2
Visualizing translation dynamics at atomic detail inside a bacterial cell
Erratum in
-
Publisher Correction: Visualizing translation dynamics at atomic detail inside a bacterial cell.Nature. 2022 Nov;611(7937):E13. doi: 10.1038/s41586-022-05455-w. Nature. 2022. PMID: 36357800 Free PMC article. No abstract available.
Abstract
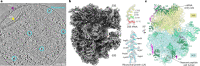
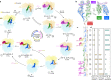
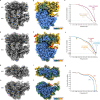
Translation is the fundamental process of protein synthesis and is catalysed by the ribosome in all living cells1. Here we use advances in cryo-electron tomography and sub-tomogram analysis2,3 to visualize the structural dynamics of translation inside the bacterium Mycoplasma pneumoniae. To interpret the functional states in detail, we first obtain a high-resolution in-cell average map of all translating ribosomes and build an atomic model for the M. pneumoniae ribosome that reveals distinct extensions of ribosomal proteins. Classification then resolves 13 ribosome states that differ in their conformation and composition. These recapitulate major states that were previously resolved in vitro, and reflect intermediates during active translation. On the basis of these states, we animate translation elongation inside native cells and show how antibiotics reshape the cellular translation landscapes. During translation elongation, ribosomes often assemble in defined three-dimensional arrangements to form polysomes4. By mapping the intracellular organization of translating ribosomes, we show that their association into polysomes involves a local coordination mechanism that is mediated by the ribosomal protein L9. We propose that an extended conformation of L9 within polysomes mitigates collisions to facilitate translation fidelity. Our work thus demonstrates the feasibility of visualizing molecular processes at atomic detail inside cells.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures














References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

