A message passing framework with multiple data integration for miRNA-disease association prediction
- PMID: 36171337
- PMCID: PMC9519928
- DOI: 10.1038/s41598-022-20529-5
A message passing framework with multiple data integration for miRNA-disease association prediction
Abstract
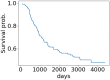
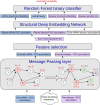
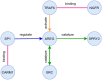
Micro RNA or miRNA is a highly conserved class of non-coding RNA that plays an important role in many diseases. Identifying miRNA-disease associations can pave the way for better clinical diagnosis and finding potential drug targets. We propose a biologically-motivated data-driven approach for the miRNA-disease association prediction, which overcomes the data scarcity problem by exploiting information from multiple data sources. The key idea is to enrich the existing miRNA/disease-protein-coding gene (PCG) associations via a message passing framework, followed by the use of disease ontology information for further feature filtering. The enriched and filtered PCG associations are then used to construct the inter-connected miRNA-PCG-disease network to train a structural deep network embedding (SDNE) model. Finally, the pre-trained embeddings and the biologically relevant features from the miRNA family and disease semantic similarity are concatenated to form the pair input representations to a Random Forest classifier whose task is to predict the miRNA-disease association probabilities. We present large-scale comparative experiments, ablation, and case studies to showcase our approach's superiority. Besides, we make the model prediction results for 1618 miRNAs and 3679 diseases, along with all related information, publicly available at http://software.mpm.leibniz-ai-lab.de/ to foster assessments and future adoption.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

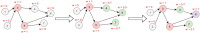

Similar articles
-
A network embedding-based multiple information integration method for the MiRNA-disease association prediction.BMC Bioinformatics. 2019 Sep 12;20(1):468. doi: 10.1186/s12859-019-3063-3. BMC Bioinformatics. 2019. PMID: 31510919 Free PMC article.
-
A structural deep network embedding model for predicting associations between miRNA and disease based on molecular association network.Sci Rep. 2021 Jun 16;11(1):12640. doi: 10.1038/s41598-021-91991-w. Sci Rep. 2021. PMID: 34135401 Free PMC article.
-
Predicting miRNA-disease association from heterogeneous information network with GraRep embedding model.Sci Rep. 2020 Apr 20;10(1):6658. doi: 10.1038/s41598-020-63735-9. Sci Rep. 2020. PMID: 32313121 Free PMC article.
-
MicroRNAs and complex diseases: from experimental results to computational models.Brief Bioinform. 2019 Mar 22;20(2):515-539. doi: 10.1093/bib/bbx130. Brief Bioinform. 2019. PMID: 29045685 Review.
-
Research progress of miRNA-disease association prediction and comparison of related algorithms.Brief Bioinform. 2022 May 13;23(3):bbac066. doi: 10.1093/bib/bbac066. Brief Bioinform. 2022. PMID: 35246678 Review.
Cited by
-
HHOMR: a hybrid high-order moment residual model for miRNA-disease association prediction.Brief Bioinform. 2024 Jul 25;25(5):bbae412. doi: 10.1093/bib/bbae412. Brief Bioinform. 2024. PMID: 39175132 Free PMC article.
-
HGCLAMIR: Hypergraph contrastive learning with attention mechanism and integrated multi-view representation for predicting miRNA-disease associations.PLoS Comput Biol. 2024 Apr 23;20(4):e1011927. doi: 10.1371/journal.pcbi.1011927. eCollection 2024 Apr. PLoS Comput Biol. 2024. PMID: 38652712 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

