Multiparameter analysis of timelapse imaging reveals kinetics of megakaryocytic erythroid progenitor clonal expansion and differentiation
- PMID: 36171423
- PMCID: PMC9519589
- DOI: 10.1038/s41598-022-19013-x
Multiparameter analysis of timelapse imaging reveals kinetics of megakaryocytic erythroid progenitor clonal expansion and differentiation
Abstract
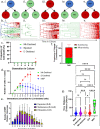
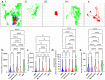
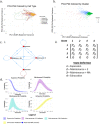
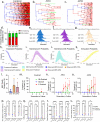
Single-cell assays have enriched our understanding of hematopoiesis and, more generally, stem and progenitor cell biology. However, these single-end-point approaches provide only a static snapshot of the state of a cell. To observe and measure dynamic changes that may instruct cell fate, we developed an approach for examining hematopoietic progenitor fate specification using long-term (> 7-day) single-cell time-lapse imaging for up to 13 generations with in situ fluorescence staining of primary human hematopoietic progenitors followed by algorithm-assisted lineage tracing. We analyzed progenitor cell dynamics, including the division rate, velocity, viability, and probability of lineage commitment at the single-cell level over time. We applied a Markov probabilistic model to predict progenitor division outcome over each generation in culture. We demonstrated the utility of this methodological pipeline by evaluating the effects of the cytokines thrombopoietin and erythropoietin on the dynamics of self-renewal and lineage specification in primary human bipotent megakaryocytic-erythroid progenitors (MEPs). Our data support the hypothesis that thrombopoietin and erythropoietin support the viability and self-renewal of MEPs, but do not affect fate specification. Thus, single-cell tracking of time-lapse imaged colony-forming unit assays provides a robust method for assessing the dynamics of progenitor self-renewal and lineage commitment.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

