Developmental dynamics of RNA translation in the human brain
- PMID: 36171426
- PMCID: PMC10198132
- DOI: 10.1038/s41593-022-01164-9
Developmental dynamics of RNA translation in the human brain
Abstract
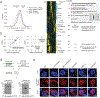
The precise regulation of gene expression is fundamental to neurodevelopment, plasticity and cognitive function. Although several studies have profiled transcription in the developing human brain, there is a gap in understanding of accompanying translational regulation. In this study, we performed ribosome profiling on 73 human prenatal and adult cortex samples. We characterized the translational regulation of annotated open reading frames (ORFs) and identified thousands of previously unknown translation events, including small ORFs that give rise to human-specific and/or brain-specific microproteins, many of which we independently verified using proteomics. Ribosome profiling in stem-cell-derived human neuronal cultures corroborated these findings and revealed that several neuronal activity-induced non-coding RNAs encode previously undescribed microproteins. Physicochemical analysis of brain microproteins identified a class of proteins that contain arginine-glycine-glycine (RGG) repeats and, thus, may be regulators of RNA metabolism. This resource expands the known translational landscape of the human brain and illuminates previously unknown brain-specific protein products.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures






References
-
- van Heesch S et al. The Translational Landscape of the Human Heart. Cell 178, 242–260.e29 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

