Oligodendrocyte precursor cells engulf synapses during circuit remodeling in mice
- PMID: 36171430
- PMCID: PMC9534756
- DOI: 10.1038/s41593-022-01170-x
Oligodendrocyte precursor cells engulf synapses during circuit remodeling in mice
Erratum in
-
Publisher Correction: Oligodendrocyte precursor cells engulf synapses during circuit remodeling in mice.Nat Neurosci. 2022 Dec;25(12):1735. doi: 10.1038/s41593-022-01209-z. Nat Neurosci. 2022. PMID: 36344700 Free PMC article. No abstract available.
Abstract
Oligodendrocyte precursor cells (OPCs) give rise to myelinating oligodendrocytes throughout life, but the functions of OPCs are not limited to oligodendrogenesis. Here we show that OPCs contribute to thalamocortical presynapse elimination in the developing and adult mouse visual cortex. OPC-mediated synapse engulfment increases in response to sensory experience during neural circuit refinement. Our data suggest that OPCs may regulate synaptic connectivity in the brain independently of oligodendrogenesis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



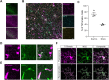
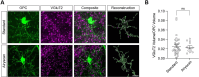




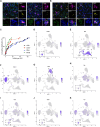


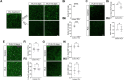
References
-
- Bergles, D. E., Roberts, J. D., Somogyi, P. & Jahr, C. E. Glutamatergic synapses on oligodendrocyte precursor cells in the hippocampus. Nature405, 187–191 (2000). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

