Assessment of small in-frame indels and C-terminal nonsense variants of BRCA1 using a validated functional assay
- PMID: 36171434
- PMCID: PMC9519549
- DOI: 10.1038/s41598-022-20500-4
Assessment of small in-frame indels and C-terminal nonsense variants of BRCA1 using a validated functional assay
Abstract
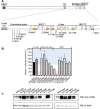
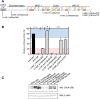
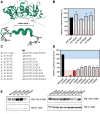
BRCA1 (Breast Cancer 1, early onset) is linked to breast and ovarian cancer predisposition. Still, the risks conferred by a significant portion of BRCA1 variants identified in the population remains unknown. Most of these variants of uncertain significance are missense alterations. However, the functional implications of small in-frame deletions and/or insertions (indels) are also difficult to predict. Our group has previously evaluated the functional impact of 347 missense variants using an extensively validated transcriptional activity assay. Here we show a systematic assessment of 30 naturally occurring in-frame indels located at the C-terminal region of BRCA1. We identified positions sensitive and tolerant to alterations, expanding the knowledge of structural determinants of BRCA1 function. We further designed and assessed the impact of four single codon deletions in the tBRCT linker region and six nonsense variants at the C-terminus end of BRCA1. Amino acid substitutions, deletions or insertions in the disordered region do not significantly impact activity and are not likely to constitute pathogenic alleles. On the other hand, a sizeable fraction of in-frame indels at the BRCT domain significantly impact function. We then use a Bayesian integrative statistical model to derive the probability of pathogenicity for each variant. Our data highlights the importance of assessing the impact of small in-frame indels in BRCA1 to improve risk assessment and clinical decisions for carriers.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Impact of amino acid substitutions at secondary structures in the BRCT domains of the tumor suppressor BRCA1: Implications for clinical annotation.J Biol Chem. 2019 Apr 12;294(15):5980-5992. doi: 10.1074/jbc.RA118.005274. Epub 2019 Feb 14. J Biol Chem. 2019. PMID: 30765603 Free PMC article.
-
Probing structure-function relationships in missense variants in the carboxy-terminal region of BRCA1.PLoS One. 2014 May 20;9(5):e97766. doi: 10.1371/journal.pone.0097766. eCollection 2014. PLoS One. 2014. PMID: 24845084 Free PMC article.
-
Comprehensive evaluation and efficient classification of BRCA1 RING domain missense substitutions.Am J Hum Genet. 2022 Jun 2;109(6):1153-1174. doi: 10.1016/j.ajhg.2022.05.004. Am J Hum Genet. 2022. PMID: 35659930 Free PMC article.
-
Understanding germ-line mutations in BRCA1.Cancer Biol Ther. 2004 Jun;3(6):515-20. doi: 10.4161/cbt.3.6.841. Epub 2004 Jun 1. Cancer Biol Ther. 2004. PMID: 15254424 Review.
-
Prevalence of specific and recurrent/founder pathogenic variants in BRCA genes in breast and ovarian cancer in North Africa.BMC Cancer. 2022 Feb 25;22(1):208. doi: 10.1186/s12885-022-09181-4. BMC Cancer. 2022. PMID: 35216584 Free PMC article.
Cited by
-
Functional analyses of rare germline BRCA1 variants by transcriptional activation and homologous recombination repair assays.BMC Cancer. 2023 Apr 21;23(1):368. doi: 10.1186/s12885-023-10790-w. BMC Cancer. 2023. PMID: 37085799 Free PMC article.
-
Functional Analyses of Rare Germline Missense BRCA1 Variants Located within and outside Protein Domains with Known Functions.Genes (Basel). 2023 Jan 19;14(2):262. doi: 10.3390/genes14020262. Genes (Basel). 2023. PMID: 36833189 Free PMC article.
-
Atypical cancer risk profile in carriers of Italian founder BRCA1 variant p.His1673del: Implications for classification and clinical management.Cancer Med. 2024 Aug;13(16):e70114. doi: 10.1002/cam4.70114. Cancer Med. 2024. PMID: 39194334 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

