Identification of a hippocampal lncRNA-regulating network in a natural aging rat model
- PMID: 36171542
- PMCID: PMC9520886
- DOI: 10.1186/s12868-022-00743-7
Identification of a hippocampal lncRNA-regulating network in a natural aging rat model
Abstract
Background: Dysregulation of long noncoding RNA (lncRNA) expression is related to aging and age-associated neurodegenerative diseases, and the lncRNA expression profile in the aging hippocampus is not well characterized. In the present investigation, the changed mRNAs and lncRNAs were confirmed via deep RNA sequencing. GO and KEGG pathway analyses were conducted to investigate the principal roles of the clearly dysregulated mRNAs and lncRNAs. Subsequently, through the prediction of miRNAs via which mRNAs and lncRNAs bind together, a competitive endogenous RNA network was constructed.
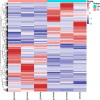
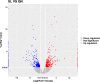
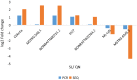
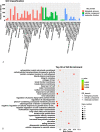
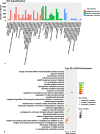
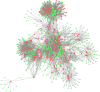
Results: A total of 447 lncRNAs and 182 mRNAs were upregulated, and 385 lncRNAs and 144 mRNAs were downregulated. Real-time reverse transcription-polymerase chain reaction validated the reliability of mRNA and lncRNA sequencing. KEGG pathway and GO analyses revealed that differentially expressed (DE) mRNAs were associated with cell adhesion molecules (CAMs), the p53 signaling pathway (SP), phagosomes, PPAR SP and ECM-receptor interactions. KEGG pathway and GO analyses showed that the target genes of the DE lncRNAs were related to cellular senescence, the p53 signaling pathway, leukocyte transendothelial migration and tyrosine metabolism. Coexpression analyses showed that 561 DE lncRNAs were associated with DE mRNAs. A total of 58 lncRNA-miRNA-mRNA target pairs were confirmed in this lncRNA‒miRNA‒mRNA network, comprising 10 mRNAs, 13 miRNAs and 38 lncRNAs.
Conclusions: We found specific lncRNAs and mRNAs in the hippocampus of natural aging model rats, as well as abnormal regulatory ceRNA networks. Our outcomes help explain the pathogenesis of brain aging and provide direction for further research.
Keywords: Aging; Hippocampus; lncRNAs; mRNAs.
© 2022. The Author(s).
Conflict of interest statement
The authors have stated no conflicts of interest.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

