Interpretation of the role of germline and somatic non-coding mutations in cancer: expression and chromatin conformation informed analysis
- PMID: 36171609
- PMCID: PMC9520844
- DOI: 10.1186/s13148-022-01342-3
Interpretation of the role of germline and somatic non-coding mutations in cancer: expression and chromatin conformation informed analysis
Abstract
Background: There has been extensive scrutiny of cancer driving mutations within the exome (especially amino acid altering mutations) as these are more likely to have a clear impact on protein functions, and thus on cell biology. However, this has come at the neglect of systematic identification of regulatory (non-coding) variants, which have recently been identified as putative somatic drivers and key germline risk factors for cancer development. Comprehensive understanding of non-coding mutations requires understanding their role in the disruption of regulatory elements, which then disrupt key biological functions such as gene expression.
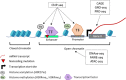
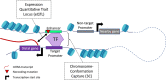
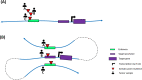
Main body: We describe how advancements in sequencing technologies have led to the identification of a large number of non-coding mutations with uncharacterized biological significance. We summarize the strategies that have been developed to interpret and prioritize the biological mechanisms impacted by non-coding mutations, focusing on recent annotation of cancer non-coding variants utilizing chromatin states, eQTLs, and chromatin conformation data.
Conclusion: We believe that a better understanding of how to apply different regulatory data types into the study of non-coding mutations will enhance the discovery of novel mechanisms driving cancer.
Keywords: Cancer; Chromosome conformation; GWAS; Germline mutation; Hi-C; Non-coding mutation; Somatic mutation; eQTL.
© 2022. The Author(s).
Conflict of interest statement
All authors have seen and approved the final manuscript. They do not have any competing interests to declare.
Figures



Similar articles
-
Hidden secrets of the cancer genome: unlocking the impact of non-coding mutations in gene regulatory elements.Cell Mol Life Sci. 2024 Jun 20;81(1):274. doi: 10.1007/s00018-024-05314-z. Cell Mol Life Sci. 2024. PMID: 38902506 Free PMC article. Review.
-
Mining the coding and non-coding genome for cancer drivers.Cancer Lett. 2015 Dec 28;369(2):307-15. doi: 10.1016/j.canlet.2015.09.015. Epub 2015 Oct 1. Cancer Lett. 2015. PMID: 26433158 Review.
-
A Comprehensive Investigation of Genomic Variants in Prostate Cancer Reveals 30 Putative Regulatory Variants.Int J Mol Sci. 2023 Jan 27;24(3):2472. doi: 10.3390/ijms24032472. Int J Mol Sci. 2023. PMID: 36768794 Free PMC article.
-
OncoBase: a platform for decoding regulatory somatic mutations in human cancers.Nucleic Acids Res. 2019 Jan 8;47(D1):D1044-D1055. doi: 10.1093/nar/gky1139. Nucleic Acids Res. 2019. PMID: 30445567 Free PMC article.
-
Candidate Cancer Driver Mutations in Distal Regulatory Elements and Long-Range Chromatin Interaction Networks.Mol Cell. 2020 Mar 19;77(6):1307-1321.e10. doi: 10.1016/j.molcel.2019.12.027. Epub 2020 Jan 17. Mol Cell. 2020. PMID: 31954095
Cited by
-
Links between melanoma germline risk loci, driver genes and comorbidities: insight from a tissue-specific multi-omic analysis.Mol Oncol. 2024 Apr;18(4):1031-1048. doi: 10.1002/1878-0261.13599. Epub 2024 Feb 3. Mol Oncol. 2024. PMID: 38308491 Free PMC article.
-
Variation within the non-coding genome influences genetic and epigenetic regulation of the human leukocyte antigen genes.Front Immunol. 2024 Sep 17;15:1422834. doi: 10.3389/fimmu.2024.1422834. eCollection 2024. Front Immunol. 2024. PMID: 39355248 Free PMC article. Review.
-
Hidden secrets of the cancer genome: unlocking the impact of non-coding mutations in gene regulatory elements.Cell Mol Life Sci. 2024 Jun 20;81(1):274. doi: 10.1007/s00018-024-05314-z. Cell Mol Life Sci. 2024. PMID: 38902506 Free PMC article. Review.
-
A Comprehensive Bioinformatics Approach to Analysis of Variants: Variant Calling, Annotation, and Prioritization.Methods Mol Biol. 2025;2889:207-233. doi: 10.1007/978-1-0716-4322-8_15. Methods Mol Biol. 2025. PMID: 39745615
-
Assessing the reliability of point mutation as data augmentation for deep learning with genomic data.BMC Bioinformatics. 2024 Apr 30;25(1):170. doi: 10.1186/s12859-024-05787-6. BMC Bioinformatics. 2024. PMID: 38689247 Free PMC article.
References
-
- Rubin CM. The genetic basis of human cancer. Ann Intern Med. 1998;129(9):759. doi: 10.7326/0003-4819-129-9-199811010-00045. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

