Selected commensals educate the intestinal vascular and immune system for immunocompetence
- PMID: 36171625
- PMCID: PMC9520927
- DOI: 10.1186/s40168-022-01353-5
Selected commensals educate the intestinal vascular and immune system for immunocompetence
Abstract
Background: The intestinal microbiota fundamentally guides the development of a normal intestinal physiology, the education, and functioning of the mucosal immune system. The Citrobacter rodentium-carrier model in germ-free (GF) mice is suitable to study the influence of selected microbes on an otherwise blunted immune response in the absence of intestinal commensals.
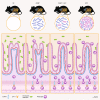
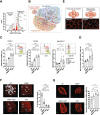
Results: Here, we describe that colonization of adult carrier mice with 14 selected commensal microbes (OMM12 + MC2) was sufficient to reestablish the host immune response to enteric pathogens; this conversion was facilitated by maturation and activation of the intestinal blood vessel system and the step- and timewise stimulation of innate and adaptive immunity. While the immature colon of C. rodentium-infected GF mice did not allow sufficient extravasation of neutrophils into the gut lumen, colonization with OMM12 + MC2 commensals initiated the expansion and activation of the visceral vascular system enabling granulocyte transmigration into the gut lumen for effective pathogen elimination.
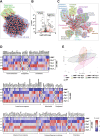
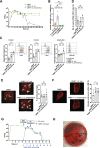
Conclusions: Consortium modeling revealed that the addition of two facultative anaerobes to the OMM12 community was essential to further progress the intestinal development. Moreover, this study demonstrates the therapeutic value of a defined consortium to promote intestinal maturation and immunity even in adult organisms. Video Abstract.
Keywords: Asymptomatic infection; Blood vessel development; C. rodentium; Colonization resistance; Commensal imprinting; Endothelial cells; Enteric pathogen; Genome-guided microbiota; Intestinal maturation; Microbial consortia; Neutrophils; Oligo-Mouse-Microbiota.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

