The Microbiome in Comedonal Contents of Inflammatory Acne Vulgaris is Composed of an Overgrowth of Cutibacterium Spp. and Other Cutaneous Microorganisms
- PMID: 36172249
- PMCID: PMC9510696
- DOI: 10.2147/CCID.S379609
The Microbiome in Comedonal Contents of Inflammatory Acne Vulgaris is Composed of an Overgrowth of Cutibacterium Spp. and Other Cutaneous Microorganisms
Abstract
Background: Acne vulgaris (acne) and cutaneous resident microorganisms are considered to be closely related. However, the bacterial and fungal microbiota in the comedonal contents of inflammatory acne lesions have not yet been investigated in detail.
Purpose: To clarify the relationship between cutaneous microorganisms and acne, we examined the microbiome in the comedonal contents of inflammatory acne and on the facial skin of patients with acne using 16s rRNA and ITS gene sequencing with a next-generation sequencer (NGS).
Patients and methods: Twenty-two untreated Japanese acne outpatients were examined. The comedonal contents of inflammatory acne lesions on the face were collected using a comedo extractor. Skin surface samples from facial skin were collected using the swab method.
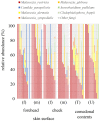
Results: The results obtained revealed that the predominant bacteria in the comedonal contents of inflammatory acne were Cutibacterium spp. (more prominent in areas with large amounts of sebum), while those on the skin surface were Staphylococcus spp. Malassezia spp., particularly Malassezia restricta, were the predominant fungi in both the comedonal contents of inflammatory acne and on the skin surface. The bacterial microbiome in comedonal contents exhibited stronger metabolic activity, including the production of enzymes related to acne, than that on the skin surface.
Conclusion: These results indicate that acne is an inflammatory disease involving the overgrowth of Cutibacterium acnes and other cutaneous resident microorganisms, including Malassezia spp.
Keywords: Cutibacterium; Malassezia; Staphylococcus; acne vulgaris; microbiome; microbiota; next generation sequencing.
© 2022 Akaza et al.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures






References
LinkOut - more resources
Full Text Sources
Miscellaneous

