Case report: Understanding the impact of persistent tissue-localization of SARS-CoV-2 on immune response activity via spatial transcriptomic analysis of two cancer patients with COVID-19 co-morbidity
- PMID: 36172383
- PMCID: PMC9510984
- DOI: 10.3389/fimmu.2022.978760
Case report: Understanding the impact of persistent tissue-localization of SARS-CoV-2 on immune response activity via spatial transcriptomic analysis of two cancer patients with COVID-19 co-morbidity
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected half a billion people, including vulnerable populations such as cancer patients. While increasing evidence supports the persistence of SARS-CoV-2 months after a negative nasopharyngeal swab test, the effects on long-term immune memory and cancer treatment are unclear. In this report, we examined post-COVID-19 tissue-localized immune responses in a hepatocellular carcinoma (HCC) patient and a colorectal cancer (CRC) patient. Using spatial whole-transcriptomic analysis, we demonstrated spatial profiles consistent with a lymphocyte-associated SARS-CoV-2 response (based on two public COVID-19 gene sets) in the tumors and adjacent normal tissues, despite intra-tumor heterogeneity. The use of RNAscope and multiplex immunohistochemistry revealed that the spatial localization of B cells was significantly associated with lymphocyte-associated SARS-CoV-2 responses within the spatial transcriptomic (ST) niches showing the highest levels of virus. Furthermore, single-cell RNA sequencing data obtained from previous (CRC) or new (HCC) ex vivo stimulation experiments showed that patient-specific SARS-CoV-2 memory B cells were the main contributors to this positive association. Finally, we evaluated the spatial associations between SARS-CoV-2-induced immunological effects and immunotherapy-related anti-tumor immune responses. Immuno-predictive scores (IMPRES) revealed consistent positive spatial correlations between T cells/cytotoxic lymphocytes and the predicted immune checkpoint blockade (ICB) response, particularly in the HCC tissues. However, the positive spatial correlation between B cells and IMPRES score was restricted to the high-virus ST niche. In addition, tumor immune dysfunction and exclusion (TIDE) analysis revealed marked T cell dysfunction and inflammation, alongside low T cell exclusion and M2 tumor-associated macrophage infiltration. Our results provide in situ evidence of SARS-CoV-2-generated persistent immunological memory, which could not only provide tissue protection against reinfection but may also modulate the tumor microenvironment, favoring ICB responsiveness. As the number of cancer patients with COVID-19 comorbidity continues to rise, improved understanding of the long-term immune response induced by SARS-CoV-2 and its impact on cancer treatment is much needed.
Keywords: SARS-CoV-2; case report; immunotherapy; intra-tumor heterogeneity; spatial transcriptomics; tissue-localized immunity.
Copyright © 2022 Lau, Yi, Goh, Cheung, Tan, Lim, Joseph, Wee, Lee, Lim, Lim, Leow, Lee, Ng, Bashiri, Cheow, Chan, Koh, Tan, Kalimuddin, Tai, Ng, Low, Lim, Liu and Yeong.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
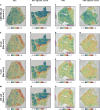
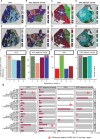


References
-
- Dai M, Liu D, Liu M, Zhou F. Patients with cancer appear more vulnerable to SARS-CoV-2: A multicenter study during the COVID-19 outbreak. Cancer Discov (2020) 10(6):783–91. doi: 10.1158/2159-8290.Cd-20-0422 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

