Aortic Cellular Diversity and Quantitative Genome-Wide Association Study Trait Prioritization Through Single-Nuclear RNA Sequencing of the Aneurysmal Human Aorta
- PMID: 36172868
- PMCID: PMC9613617
- DOI: 10.1161/ATVBAHA.122.317953
Aortic Cellular Diversity and Quantitative Genome-Wide Association Study Trait Prioritization Through Single-Nuclear RNA Sequencing of the Aneurysmal Human Aorta
Abstract
Background: Mural cells in ascending aortic aneurysms undergo phenotypic changes that promote extracellular matrix destruction and structural weakening. To explore this biology, we analyzed the transcriptional features of thoracic aortic tissue.
Methods: Single-nuclear RNA sequencing was performed on 13 samples from human donors, 6 with thoracic aortic aneurysm, and 7 without aneurysm. Individual transcriptomes were then clustered based on transcriptional profiles. Clusters were used for between-disease differential gene expression analyses, subcluster analysis, and analyzed for intersection with genetic aortic trait data.
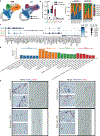
Results: We sequenced 71 689 nuclei from human thoracic aortas and identified 14 clusters, aligning with 11 cell types, predominantly vascular smooth muscle cells (VSMCs) consistent with aortic histology. With unbiased methodology, we found 7 vascular smooth muscle cell and 6 fibroblast subclusters. Differentially expressed genes analysis revealed a vascular smooth muscle cell group accounting for the majority of differential gene expression. Fibroblast populations in aneurysm exhibit distinct behavior with almost complete disappearance of quiescent fibroblasts. Differentially expressed genes were used to prioritize genes at aortic diameter and distensibility genome-wide association study loci highlighting the genes JUN, LTBP4 (latent transforming growth factor beta-binding protein 1), and IL34 (interleukin 34) in fibroblasts, ENTPD1, PDLIM5 (PDZ and LIM domain 5), ACTN4 (alpha-actinin-4), and GLRX in vascular smooth muscle cells, as well as LRP1 in macrophage populations.
Conclusions: Using nuclear RNA sequencing, we describe the cellular diversity of healthy and aneurysmal human ascending aorta. Sporadic aortic aneurysm is characterized by differential gene expression within known cellular classes rather than by the appearance of novel cellular forms. Single-nuclear RNA sequencing of aortic tissue can be used to prioritize genes at aortic trait loci.
Keywords: aortic aneurysms; extracellular matrix; fibroblast; genome-wide association study; thoracic; transcriptomes.
Figures

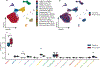




Comment in
-
New Genomic Techniques Provide Novel Insights Into Ascending Aortic Aneurysm Pathology.Arterioscler Thromb Vasc Biol. 2022 Nov;42(11):1375-1377. doi: 10.1161/ATVBAHA.122.318525. Epub 2022 Oct 6. Arterioscler Thromb Vasc Biol. 2022. PMID: 36200445 No abstract available.
References
-
- Vilacosta I, San Román JA, di Bartolomeo R, Eagle K, Estrera AL, Ferrera C, Kaji S, Nienaber CA, Riambau V, Schäfers HJ, Serrano FJ, Song JK, Maroto L. Acute Aortic Syndrome Revisited: JACC State-of-the-Art Review. J Am Coll Cardiol. 2021;78(21):2106–2125. doi: 10.1016/j.jacc.2021.09.022 - DOI - PubMed
-
- Erbel R, Aboyans V, Boileau C, Bossone E, Bartolomeo RD, Eggebrecht H, Evangelista A, Falk V, Frank H, Gaemperli O, Grabenwöger M, Haverich A, Iung B, Manolis AJ, Meijboom F, Nienaber CA, Roffi M, Rousseau H, Sechtem U, Sirnes PA, Allmen RS von, Vrints CJM, ESC Committee for Practice Guidelines. 2014 ESC Guidelines on the diagnosis and treatment of aortic diseases: Document covering acute and chronic aortic diseases of the thoracic and abdominal aorta of the adult. The Task Force for the Diagnosis and Treatment of Aortic Diseases of the European Society of Cardiology (ESC). Eur Heart J. 2014;35(41):2873–2926. doi: 10.1093/eurheartj/ehu281 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

