CBP-Mediated Acetylation of Importin α Mediates Calcium-Dependent Nucleocytoplasmic Transport of Selective Proteins in Drosophila Neurons
- PMID: 36172977
- PMCID: PMC9676984
- DOI: 10.14348/molcells.2022.0104
CBP-Mediated Acetylation of Importin α Mediates Calcium-Dependent Nucleocytoplasmic Transport of Selective Proteins in Drosophila Neurons
Abstract
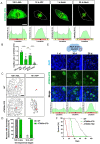
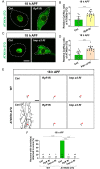
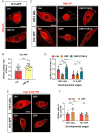
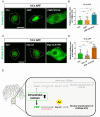
For proper function of proteins, their subcellular localization needs to be monitored and regulated in response to the changes in cellular demands. In this regard, dysregulation in the nucleocytoplasmic transport (NCT) of proteins is closely associated with the pathogenesis of various neurodegenerative diseases. However, it remains unclear whether there exists an intrinsic regulatory pathway(s) that controls NCT of proteins either in a commonly shared manner or in a target-selectively different manner. To dissect between these possibilities, in the current study, we investigated the molecular mechanism regulating NCT of truncated ataxin-3 (ATXN3) proteins of which genetic mutation leads to a type of polyglutamine (polyQ) diseases, in comparison with that of TDP-43. In Drosophila dendritic arborization (da) neurons, we observed dynamic changes in the subcellular localization of truncated ATXN3 proteins between the nucleus and the cytosol during development. Moreover, ectopic neuronal toxicity was induced by truncated ATXN3 proteins upon their nuclear accumulation. Consistent with a previous study showing intracellular calcium-dependent NCT of TDP-43, NCT of ATXN3 was also regulated by intracellular calcium level and involves Importin α3 (Imp α3). Interestingly, NCT of ATXN3, but not TDP-43, was primarily mediated by CBP. We further showed that acetyltransferase activity of CBP is important for NCT of ATXN3, which may acetylate Imp α3 to regulate NCT of ATXN3. These findings demonstrate that CBP-dependent acetylation of Imp α3 is crucial for intracellular calcium-dependent NCT of ATXN3 proteins, different from that of TDP-43, in Drosophila neurons.
Keywords: ATXN3; CBP; Importin α; acetylation; calcium; nucleocytoplasmic transport.
Conflict of interest statement
The authors have no potential conflicts of interest to disclose.
Figures

Similar articles
-
Cytosolic calcium regulates cytoplasmic accumulation of TDP-43 through Calpain-A and Importin α3.Elife. 2020 Dec 11;9:e60132. doi: 10.7554/eLife.60132. Elife. 2020. PMID: 33305734 Free PMC article.
-
Karyopherin α-3 is a key protein in the pathogenesis of spinocerebellar ataxia type 3 controlling the nuclear localization of ataxin-3.Proc Natl Acad Sci U S A. 2018 Mar 13;115(11):E2624-E2633. doi: 10.1073/pnas.1716071115. Epub 2018 Feb 23. Proc Natl Acad Sci U S A. 2018. PMID: 29476013 Free PMC article.
-
A fine balance between Prpf19 and Exoc7 in achieving degradation of aggregated protein and suppression of cell death in spinocerebellar ataxia type 3.Cell Death Dis. 2021 Feb 2;12(2):136. doi: 10.1038/s41419-021-03444-x. Cell Death Dis. 2021. PMID: 33542212 Free PMC article.
-
Importin α: a key molecule in nuclear transport and non-transport functions.J Biochem. 2016 Aug;160(2):69-75. doi: 10.1093/jb/mvw036. Epub 2016 Jun 11. J Biochem. 2016. PMID: 27289017 Review.
-
Importin α: functions as a nuclear transport factor and beyond.Proc Jpn Acad Ser B Phys Biol Sci. 2018;94(7):259-274. doi: 10.2183/pjab.94.018. Proc Jpn Acad Ser B Phys Biol Sci. 2018. PMID: 30078827 Free PMC article. Review.
Cited by
-
A comparison study of pathological features and drug efficacy between Drosophila models of C9orf72 ALS/FTD.Mol Cells. 2024 Jan;47(1):100005. doi: 10.1016/j.mocell.2023.12.003. Epub 2023 Dec 15. Mol Cells. 2024. PMID: 38376483 Free PMC article.
-
Unveiling the complexity: assessing models describing the structure and function of the nuclear pore complex.Front Cell Dev Biol. 2023 Oct 9;11:1245939. doi: 10.3389/fcell.2023.1245939. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37876551 Free PMC article. Review.
-
Drosophila Importin Alpha 1 (Dα1) Is Required to Maintain Germline Stem Cells in the Testis Niche.Cells. 2024 Mar 12;13(6):494. doi: 10.3390/cells13060494. Cells. 2024. PMID: 38534338 Free PMC article.
References
-
- Bichelmeier U., Schmidt T., Hubener J., Boy J., Ruttiger L., Habig K., Poths S., Bonin M., Knipper M., Schmidt W.J., et al. Nuclear localization of ataxin-3 is required for the manifestation of symptoms in SCA3: in vivo evidence. J. Neurosci. 2007;27:7418–7428. doi: 10.1523/JNEUROSCI.4540-06.2007. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

