Effects of targeting signal mutations in a mitochondrial presequence on the spatial distribution of the conformational ensemble in the binding site of Tom20
- PMID: 36173160
- PMCID: PMC9490799
- DOI: 10.1002/pro.4433
Effects of targeting signal mutations in a mitochondrial presequence on the spatial distribution of the conformational ensemble in the binding site of Tom20
Abstract
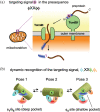
The 20-kDa TOM (translocase of outer mitochondrial membrane) subunit, Tom20, is the first receptor of the protein import pathway into mitochondria. Tom20 recognizes the mitochondrial targeting signal embedded in the presequences attached to mature mitochondrial proteins, as an N-terminal extension. Consequently, ~1,000 different mitochondrial proteins are sorted into the mitochondrial matrix, and distinguished from non-mitochondrial proteins. We previously reported the MPRIDE (multiple partial recognitions in dynamic equilibrium) mechanism to explain the structural basis of the promiscuous recognition of presequences by Tom20. A subset of the targeting signal features is recognized in each pose of the presequence in the binding state, and all of the features are collectively recognized in the dynamic equilibrium between the poses. Here, we changed the volumes of the hydrophobic side chains in the targeting signal, while maintaining the binding affinity. We tethered the mutated presequences to the binding site of Tom20 and placed them in the crystal contact-free space (CCFS) created in the crystal lattice. The spatial distributions of the mutated presequences were visualized as smeared electron densities in the low-pass filtered difference maps obtained by X-ray crystallography. The mutated presequence ensembles shifted their positions in the binding state to accommodate the larger side chains, thus providing positive evidence supporting the use of the MPRIDE mechanism in the promiscuous recognition by Tom20.
Keywords: Tom20; crystal contact-free space; mitochondrial targeting signal; presequence; promiscuous recognition.
© 2022 The Protein Society.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Wiedemann N, Pfanner N. Mitochondrial machineries for protein import and assembly. Annu Rev Biochem. 2017;86:685–714. - PubMed
-
- Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. - PubMed
-
- Araiso Y, Imai K, Endo T. Role of the TOM complex in protein import into mitochondria: Structural views. Annu Rev Biochem. 2022;91:1–25. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

