PDBe and PDBe-KB: Providing high-quality, up-to-date and integrated resources of macromolecular structures to support basic and applied research and education
- PMID: 36173162
- PMCID: PMC9517934
- DOI: 10.1002/pro.4439
PDBe and PDBe-KB: Providing high-quality, up-to-date and integrated resources of macromolecular structures to support basic and applied research and education
Abstract
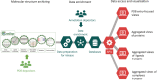
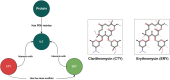
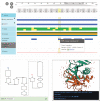
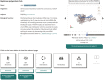
The archiving and dissemination of protein and nucleic acid structures as well as their structural, functional and biophysical annotations is an essential task that enables the broader scientific community to conduct impactful research in multiple fields of the life sciences. The Protein Data Bank in Europe (PDBe; pdbe.org) team develops and maintains several databases and web services to address this fundamental need. From data archiving as a member of the Worldwide PDB consortium (wwPDB; wwpdb.org), to the PDBe Knowledge Base (PDBe-KB; pdbekb.org), we provide data, data-access mechanisms, and visualizations that facilitate basic and applied research and education across the life sciences. Here, we provide an overview of the structural data and annotations that we integrate and make freely available. We describe the web services and data visualization tools we offer, and provide information on how to effectively use or even further develop them. Finally, we discuss the direction of our data services, and how we aim to tackle new challenges that arise from the recent, unprecedented advances in the field of structure determination and protein structure modeling.
Keywords: bioinformatics; databases; protein data bank; structural biology.
© 2022 The Authors. Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
- BB/R015201/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 223739/Z/21/Z/WT_/Wellcome Trust/United Kingdom
- BB/V004247/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/V016113/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/S017135/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 221327/Z/20/Z/WT_/Wellcome Trust/United Kingdom
- BB/T01959X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/P025846/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M011674/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/P024351/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 104948/Z/14/Z/WT_/Wellcome Trust/United Kingdom
- 218303/Z/19/Z/WT_/Wellcome Trust/United Kingdom
- BB/W017970/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/V018779/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/S020071/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources

