yEvo: experimental evolution in high school classrooms selects for novel mutations that impact clotrimazole resistance in Saccharomyces cerevisiae
- PMID: 36173330
- PMCID: PMC9635649
- DOI: 10.1093/g3journal/jkac246
yEvo: experimental evolution in high school classrooms selects for novel mutations that impact clotrimazole resistance in Saccharomyces cerevisiae
Abstract
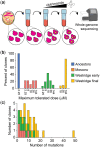
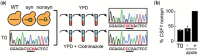
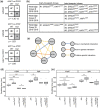
Antifungal resistance in pathogenic fungi is a growing global health concern. Nonpathogenic laboratory strains of Saccharomyces cerevisiae are an important model for studying mechanisms of antifungal resistance that are relevant to understanding the same processes in pathogenic fungi. We have developed a series of laboratory modules in which high school students used experimental evolution to study antifungal resistance by isolating azole-resistant S. cerevisiae mutants and examining the genetic basis of resistance. We have sequenced 99 clones from these experiments and found that all possessed mutations previously shown to impact azole resistance, validating our approach. We additionally found recurrent mutations in an mRNA degradation pathway and an uncharacterized mitochondrial protein (Csf1) that have possible mechanistic connections to azole resistance. The scale of replication in this initiative allowed us to identify candidate epistatic interactions, as evidenced by pairs of mutations that occur in the same clone more frequently than expected by chance (positive epistasis) or less frequently (negative epistasis). We validated one of these pairs, a negative epistatic interaction between gain-of-function mutations in the multidrug resistance transcription factors Pdr1 and Pdr3. This high school-university collaboration can serve as a model for involving members of the broader public in the scientific process to make meaningful discoveries in biomedical research.
Keywords: azole resistance; course-based research experience; experimental evolution; genome sequencing; science education; yeast.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America.
Figures

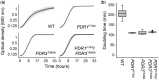
References
-
- Allen D, Wilson D, Drew R, Perfect J.. Azole antifungals: 35 years of invasive fungal infection management. Expert Rev Anti-Infect Ther. 2015;13(6):787–798. - PubMed
-
- Balzi E, Chen W, Ulaszewski S, Capieaux E, Goffeau A.. The multidrug resistance gene PDR1 from Saccharomyces cerevisiae. J Biol Chem. 1987;262(35):16871–16879. - PubMed
-
- Balzi E, Goffeau A.. Multiple or pleiotropic drug resistance in yeast. Biochim Biophys Acta. 1991;1073(2):241–252. - PubMed
-
- Baudry K, Swain E, Rahier A, Germann M, Batta A, Rondet S, Mandala S, Henry K, Tint GS, Edlind T, et al. The effect of the erg26-1 mutation on the regulation of lipid metabolism in Saccharomyces cerevisiae. J Biol Chem. 2001;276(16):12702–12711. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
