Recurrent but Short-Lived Duplications of Centromeric Proteins in Holocentric Caenorhabditis Species
- PMID: 36173809
- PMCID: PMC9577544
- DOI: 10.1093/molbev/msac206
Recurrent but Short-Lived Duplications of Centromeric Proteins in Holocentric Caenorhabditis Species
Abstract
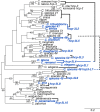
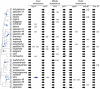
Centromeric histones (CenH3s) are essential for chromosome inheritance during cell division in most eukaryotes. CenH3 genes have rapidly evolved and undergone repeated gene duplications and diversification in many plant and animal species. In Caenorhabditis species, two independent duplications of CenH3 (named hcp-3 for HoloCentric chromosome-binding Protein 3) were previously identified in C. elegans and C. remanei. Using phylogenomic analyses in 32 Caenorhabditis species, we find strict retention of the ancestral hcp-3 gene and 10 independent duplications. Most hcp-3L (hcp-3-like) paralogs are only found in 1-2 species, are expressed in both males and females/hermaphrodites, and encode histone fold domains with 69-100% identity to ancestral hcp-3. We identified novel N-terminal protein motifs, including putative kinetochore protein-interacting motifs and a potential separase cleavage site, which are well conserved across Caenorhabditis HCP-3 proteins. Other N-terminal motifs vary in their retention across paralogs or species, revealing potential subfunctionalization or functional loss following duplication. An N-terminal extension in the hcp-3L gene of C. afra revealed an unprecedented protein fusion, where hcp-3L fused to duplicated segments from hcp-4 (nematode CENP-C). By extending our analyses beyond CenH3, we found gene duplications of six inner and outer kinetochore genes in Caenorhabditis, which appear to have been retained independent of hcp-3 duplications. Our findings suggest that centromeric protein duplications occur frequently in Caenorhabditis nematodes, are selectively retained for short evolutionary periods, then degenerate or are lost entirely. We hypothesize that unique challenges associated with holocentricity in Caenorhabditis may lead to this rapid "revolving door" of kinetochore protein paralogs.
Keywords: centromeric histone; gene duplication; kinetochore; protein motifs.
© The Author(s) 2022. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures




Similar articles
-
Separase Cleaves the N-Tail of the CENP-A Related Protein CPAR-1 at the Meiosis I Metaphase-Anaphase Transition in C. elegans.PLoS One. 2015 Apr 28;10(4):e0125382. doi: 10.1371/journal.pone.0125382. eCollection 2015. PLoS One. 2015. PMID: 25919583 Free PMC article.
-
HCP-1, a protein involved in chromosome segregation, is localized to the centromere of mitotic chromosomes in Caenorhabditis elegans.J Cell Biol. 1999 Nov 1;147(3):471-80. doi: 10.1083/jcb.147.3.471. J Cell Biol. 1999. PMID: 10545493 Free PMC article.
-
Evidence for centromere drive in the holocentric chromosomes of Caenorhabditis.PLoS One. 2012;7(1):e30496. doi: 10.1371/journal.pone.0030496. Epub 2012 Jan 23. PLoS One. 2012. PMID: 22291967 Free PMC article.
-
"Holo"er than thou: chromosome segregation and kinetochore function in C. elegans.Chromosome Res. 2004;12(6):641-53. doi: 10.1023/B:CHRO.0000036588.42225.2f. Chromosome Res. 2004. PMID: 15289669 Review.
-
"Lessons from the extremes: Epigenetic and genetic regulation in point monocentromere and holocentromere establishment on artificial chromosomes".Exp Cell Res. 2020 May 15;390(2):111974. doi: 10.1016/j.yexcr.2020.111974. Epub 2020 Mar 26. Exp Cell Res. 2020. PMID: 32222413 Review.
Cited by
-
Regulation of outer kinetochore assembly during meiosis I and II by CENP-A and KNL-2/M18BP1 in C. elegans oocytes.Curr Biol. 2024 Nov 4;34(21):4853-4868.e6. doi: 10.1016/j.cub.2024.09.004. Epub 2024 Sep 30. Curr Biol. 2024. PMID: 39353426
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

