Oncometabolite d-2HG alters T cell metabolism to impair CD8+ T cell function
- PMID: 36173860
- PMCID: PMC9629749
- DOI: 10.1126/science.abj5104
Oncometabolite d-2HG alters T cell metabolism to impair CD8+ T cell function
Abstract
Gain-of-function mutations in isocitrate dehydrogenase (IDH) in human cancers result in the production of d-2-hydroxyglutarate (d-2HG), an oncometabolite that promotes tumorigenesis through epigenetic alterations. The cancer cell-intrinsic effects of d-2HG are well understood, but its tumor cell-nonautonomous roles remain poorly explored. We compared the oncometabolite d-2HG with its enantiomer, l-2HG, and found that tumor-derived d-2HG was taken up by CD8+ T cells and altered their metabolism and antitumor functions in an acute and reversible fashion. We identified the glycolytic enzyme lactate dehydrogenase (LDH) as a molecular target of d-2HG. d-2HG and inhibition of LDH drive a metabolic program and immune CD8+ T cell signature marked by decreased cytotoxicity and impaired interferon-γ signaling that was recapitulated in clinical samples from human patients with IDH1 mutant gliomas.
Conflict of interest statement
Figures
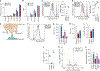
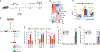
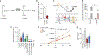



Comment in
-
Metabolite-driven antitumor immunity.Science. 2022 Sep 30;377(6614):1488-1489. doi: 10.1126/science.ade3697. Epub 2022 Sep 29. Science. 2022. PMID: 36173838
-
Tumour-derived D-2HG blocks T cell cytotoxicity.Nat Rev Cancer. 2022 Dec;22(12):657. doi: 10.1038/s41568-022-00534-6. Nat Rev Cancer. 2022. PMID: 36376756 No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

