A Novel MIP-1-Expressing Macrophage Subtype in BAL Fluid from Healthy Volunteers
- PMID: 36174229
- PMCID: PMC9986555
- DOI: 10.1165/rcmb.2021-0123OC
A Novel MIP-1-Expressing Macrophage Subtype in BAL Fluid from Healthy Volunteers
Abstract
Tissue availability remains an important limitation of single-cell genomic technologies for investigating cellular heterogeneity in human health and disease. BAL represents a minimally invasive approach to assessing an individual's lung cellular environment for diagnosis and research. However, the lack of high-quality, healthy lung reference data is a major obstacle to using single-cell approaches to study a plethora of lung diseases. Here, we performed single-cell RNA sequencing on over 40,000 cells isolated from the BAL of four healthy volunteers. Of the six cell types or lineages we identified, macrophages were consistently the most numerous across individuals. Our analysis confirmed the expression of marker genes defining cell types despite background signals because of the ambient RNA found in many single-cell studies. We assessed the variability of gene expression across macrophages and defined a distinct subpopulation of cells expressing a set of genes associated with Macrophage Inflammatory Protein 1 (MIP-1). RNA in situ hybridization and reanalysis of published lung single-cell data validated the presence of this macrophage subpopulation. Thus, our study characterizes lung macrophage heterogeneity in healthy individuals and provides a valuable resource for future studies to understand the lung environment in health and disease.
Keywords: BAL; genomics; heterogeneity; lung immunology; macrophage.
Figures
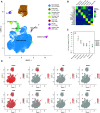
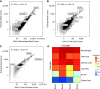
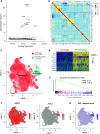
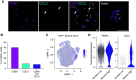
References
-
- Franks TJ, Colby TV, Travis WD, Tuder RM, Reynolds HY, Brody AR, et al. Resident cellular components of the human lung: current knowledge and goals for research on cell phenotyping and function. Proc Am Thorac Soc . 2008;5:763–766. - PubMed
-
- Agostini C, Chilosi M, Zambello R, Trentin L, Semenzato G. Pulmonary immune cells in health and disease: lymphocytes. Eur Respir J . 1993;6:1378–1401. - PubMed
-
- Hussell T, Bell TJ. Alveolar macrophages: plasticity in a tissue-specific context. Nat Rev Immunol . 2014;14:81–93. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

