Bacteriophage-antibiotic combination therapy against extensively drug-resistant Pseudomonas aeruginosa infection to allow liver transplantation in a toddler
- PMID: 36175406
- PMCID: PMC9523064
- DOI: 10.1038/s41467-022-33294-w
Bacteriophage-antibiotic combination therapy against extensively drug-resistant Pseudomonas aeruginosa infection to allow liver transplantation in a toddler
Abstract
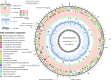
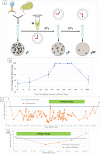
Post-operative bacterial infections are a leading cause of mortality and morbidity after ongoing liver transplantation. Bacteria causing these infections in the hospital setting can exhibit high degrees of resistance to multiple types of antibiotics, which leads to major therapeutic hurdles. Alternate ways of treating these antibiotic-resistant infections are thus urgently needed. Phage therapy is one of them and consists in using selected bacteriophage viruses - viruses who specifically prey on bacteria, naturally found in various environmental samples - as bactericidal agents in replacement or in combination with antibiotics. The use of phage therapy raises various research questions to further characterize what determines therapeutic success or failure. In this work, we report the story of a toddler who suffered from extensively drug-resistant Pseudomonas aeruginosa sepsis after liver transplantation. He was treated by a bacteriophage-antibiotic intravenous combination therapy for 86 days. This salvage therapy was well tolerated, without antibody-mediated phage neutralization. It was associated with objective clinical and microbiological improvement, eventually allowing for liver retransplantation and complete resolution of all infections. Clear in vitro phage-antibiotic synergies were observed. The occurrence of bacterial phage resistance did not result in therapeutic failure, possibly due to phage-induced virulence tradeoffs, which we investigated in different experimental models.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

