Genomic basis of insularity and ecological divergence in barn owls (Tyto alba) of the Canary Islands
- PMID: 36175501
- PMCID: PMC9613907
- DOI: 10.1038/s41437-022-00562-w
Genomic basis of insularity and ecological divergence in barn owls (Tyto alba) of the Canary Islands
Abstract
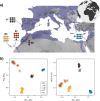
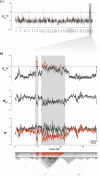
Islands, and the particular organisms that populate them, have long fascinated biologists. Due to their isolation, islands offer unique opportunities to study the effect of neutral and adaptive mechanisms in determining genomic and phenotypical divergence. In the Canary Islands, an archipelago rich in endemics, the barn owl (Tyto alba), present in all the islands, is thought to have diverged into a subspecies (T. a. gracilirostris) on the eastern ones, Fuerteventura and Lanzarote. Taking advantage of 40 whole-genomes and modern population genomics tools, we provide the first look at the origin and genetic makeup of barn owls of this archipelago. We show that the Canaries hold diverse, long-standing and monophyletic populations with a neat distinction of gene pools from the different islands. Using a new method, less sensitive to structure than classical FST, to detect regions involved in local adaptation to insular environments, we identified a haplotype-like region likely under selection in all Canaries individuals and genes in this region suggest morphological adaptations to insularity. In the eastern islands, where the subspecies is present, genomic traces of selection pinpoint signs of adapted body proportions and blood pressure, consistent with the smaller size of this population living in a hot arid climate. In turn, genomic regions under selection in the western barn owls from Tenerife showed an enrichment in genes linked to hypoxia, a potential response to inhabiting a small island with a marked altitudinal gradient. Our results illustrate the interplay of neutral and adaptive forces in shaping divergence and early onset speciation.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Insularity and the evolution of melanism, sexual dichromatism and body size in the worldwide-distributed barn owl.J Evol Biol. 2010 May;23(5):925-34. doi: 10.1111/j.1420-9101.2010.01961.x. Epub 2010 Mar 5. J Evol Biol. 2010. PMID: 20298444
-
Avian Binocularity and Adaptation to Nocturnal Environments: Genomic Insights from a Highly Derived Visual Phenotype.Genome Biol Evol. 2019 Aug 1;11(8):2244-2255. doi: 10.1093/gbe/evz111. Genome Biol Evol. 2019. PMID: 31386143 Free PMC article.
-
Genomic consequences of colonisation, migration and genetic drift in barn owl insular populations of the eastern Mediterranean.Mol Ecol. 2022 Mar;31(5):1375-1388. doi: 10.1111/mec.16324. Epub 2021 Dec 23. Mol Ecol. 2022. PMID: 34894026 Free PMC article.
-
Comprehensive molecular phylogeny of barn owls and relatives (Family: Tytonidae), and their six major Pleistocene radiations.Mol Phylogenet Evol. 2018 Aug;125:127-137. doi: 10.1016/j.ympev.2018.03.013. Epub 2018 Mar 11. Mol Phylogenet Evol. 2018. PMID: 29535030
-
Divergent selection and heterogeneous genomic divergence.Mol Ecol. 2009 Feb;18(3):375-402. doi: 10.1111/j.1365-294X.2008.03946.x. Epub 2008 Dec 29. Mol Ecol. 2009. PMID: 19143936 Review.
Cited by
-
Too big to purge: persistence of deleterious Mutations in Island populations of the European Barn Owl (Tyto alba).Heredity (Edinb). 2024 Dec;133(6):437-449. doi: 10.1038/s41437-024-00728-8. Epub 2024 Oct 13. Heredity (Edinb). 2024. PMID: 39397112 Free PMC article.
-
The recombination landscape of the barn owl, from families to populations.Genetics. 2025 Jan 8;229(1):1-50. doi: 10.1093/genetics/iyae190. Genetics. 2025. PMID: 39545468 Free PMC article.
-
The Relative Roles of Selection and Drift in the Chaffinch Radiation (Aves: Fringilla) Across the Atlantic Archipelagos of Macaronesia.Ecol Evol. 2025 Apr 15;15(4):e71307. doi: 10.1002/ece3.71307. eCollection 2025 Apr. Ecol Evol. 2025. PMID: 40242797 Free PMC article.
-
An allele-sharing, moment-based estimator of global, population-specific and population-pair FST under a general model of population structure.PLoS Genet. 2023 Nov 27;19(11):e1010871. doi: 10.1371/journal.pgen.1010871. eCollection 2023 Nov. PLoS Genet. 2023. PMID: 38011288 Free PMC article. Review.
References
-
- Cumer T, Machado AP, Dumont G, Bontzorlos VA, Ceccherelli R, Charter M, Dichmann K, Martens H-D, Kassinis N, Lourenço R, Manzia F, Ovari K, Prévost L, Rakovic M, Siverio F, Roulin A, and Goudet J (2021) Population genomics of barn owls in the Western Parlearctic; NCBI bio project PRJNA727977; 10.1093/molbev/msab343
-
- Machado AP, Cumer T, Iseli C, Beaudoing E, Dupasquier M, Guex N, Dichmann K, Lourenço R, Lusby J, Martens H-D, Prévost L, Ramsden D, Roulin A, and Goudet J (2021) Population genomics of barn owls in the British Isles; NCBI bio project PRJNA700797; 10.1111/mec.16250
-
- Anguita F and Hernán F (2000) The Canary Islands origin: A unifying model. J Volcanol Geotherm Res 103:1–26. Elsevier B.V
-
- Astle WJ, Elding H, Jiang T, Allen D, Ruklisa D, Mann AL, Mead D, Bouman H, Riveros-Mckay F, Kostadima MA, Lambourne JJ, Sivapalaratnam S, Downes K, Kundu K, Bomba L, Berentsen K, Bradley JR, Daugherty LC, Delaneau O, Freson K, Garner SF, Grassi L, Guerrero J, Haimel M, Janssen-Megens EM, Kaan A, Kamat M, Kim B, Mandoli A, Marchini J, Martens JHA, Meacham S, Megy K, O’Connell J, Petersen R, Sharifi N, Sheard SM, Staley JR, Tuna S, van der Ent M, Walter K, Wang SY, Wheeler E, Wilder SP, Iotchkova V, Moore C, Sambrook J, Stunnenberg HG, Di Angelantonio E, Kaptoge S, Kuijpers TW, Carrillo-de-Santa-Pau E, Juan D, Rico D, Valencia A, Chen L, Ge B, Vasquez L, Kwan T, Garrido-Martín D, Watt S, Yang Y, Guigo R, Beck S, Paul DS, Pastinen T, Bujold D, Bourque G, Frontini M, Danesh J, Roberts DJ, Ouwehand WH, Butterworth AS, Soranzo N. The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell. 2016;167:1415–1429.e19. - PMC - PubMed
-
- Balloux F. Heterozygote excess in small populations and the heterozygote-excess effective population size. Evolution. 2004;58:1891–1900. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

