Divergent evolutionary trajectories of bryophytes and tracheophytes from a complex common ancestor of land plants
- PMID: 36175544
- PMCID: PMC9630106
- DOI: 10.1038/s41559-022-01885-x
Divergent evolutionary trajectories of bryophytes and tracheophytes from a complex common ancestor of land plants
Abstract
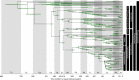
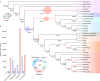
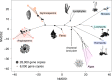
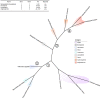
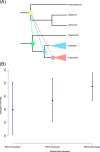
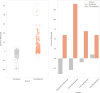
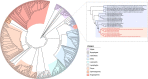
The origin of plants and their colonization of land fundamentally transformed the terrestrial environment. Here we elucidate the basis of this formative episode in Earth history through patterns of lineage, gene and genome evolution. We use new fossil calibrations, a relative clade age calibration (informed by horizontal gene transfer) and new phylogenomic methods for mapping gene family origins. Distinct rooting strategies resolve tracheophytes (vascular plants) and bryophytes (non-vascular plants) as monophyletic sister groups that diverged during the Cambrian, 515-494 million years ago. The embryophyte stem is characterized by a burst of gene innovation, while bryophytes subsequently experienced an equally dramatic episode of reductive genome evolution in which they lost genes associated with the elaboration of vasculature and the stomatal complex. Overall, our analyses reveal that extant tracheophytes and bryophytes are both highly derived from a more complex ancestral land plant. Understanding the origin of land plants requires tracing character evolution across a diversity of modern lineages.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

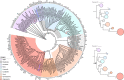


References
-
- Berry, J. A., Beerling, D. J. & Franks, P. J. Stomata: key players in the Earth system, past and present. Curr. Opin. Plant Biol.10.1016/j.pbi.2010.04.013 (2010). - PubMed
-
- Wellman CH, Strother PK. The terrestrial biota prior to the origin of land plants (embryophytes): a review of the evidence. Palaeontology. 2015;58:601–627.
-
- Christenhusz, M. J. M. & Byng, J. W. The number of known plants species in the world and its annual increase. Phytotaxa10.11646/phytotaxa.261.3.1 (2016).
-
- de Vries, J. & Archibald, J. M. Plant evolution: landmarks on the path to terrestrial life. N. Phytol. 10.1111/nph.14975 (2018). - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources

