Targeted protein S-nitrosylation of ACE2 inhibits SARS-CoV-2 infection
- PMID: 36175661
- PMCID: PMC10127945
- DOI: 10.1038/s41589-022-01149-6
Targeted protein S-nitrosylation of ACE2 inhibits SARS-CoV-2 infection
Abstract
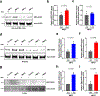
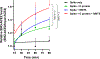
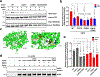
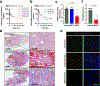
Prevention of infection and propagation of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a high priority in the Coronavirus Disease 2019 (COVID-19) pandemic. Here we describe S-nitrosylation of multiple proteins involved in SARS-CoV-2 infection, including angiotensin-converting enzyme 2 (ACE2), the receptor for viral entry. This reaction prevents binding of ACE2 to the SARS-CoV-2 spike protein, thereby inhibiting viral entry, infectivity and cytotoxicity. Aminoadamantane compounds also inhibit coronavirus ion channels formed by envelope (E) protein. Accordingly, we developed dual-mechanism aminoadamantane nitrate compounds that inhibit viral entry and, thus, the spread of infection by S-nitrosylating ACE2 via targeted delivery of the drug after E protein channel blockade. These non-toxic compounds are active in vitro and in vivo in the Syrian hamster COVID-19 model and, thus, provide a novel avenue to pursue therapy.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
The authors declare that S.A.L. is an inventor on patents for the use of memantine and various aminoadamantane nitrate compounds for neurodegenerative and neurodevelopmental disorders. He is also an inventor on composition of matter patents and use patents for aminoadamantane nitrate compounds in treating COVID-19 and other viral diseases. Per Harvard University guidelines, S.A.L. participates in a royalty-sharing agreement with his former institution Boston Children’s Hospital/Harvard Medical School, which licensed the drug memantine (Namenda®) to Forest Laboratories, Inc./Actavis/Allergan/AbbVie for use in dementia. The aminoadamantane nitrate compounds have been licensed to EuMentis Therapeutics, Inc. (Newton, Massachusetts). C.B. is a chemist employed at EuMentis Therapeutics. The other authors declare no financial conflicts of interest relevant to this publication.
Figures












Update of
-
Targeted protein S-nitrosylation of ACE2 as potential treatment to prevent spread of SARS-CoV-2 infection.bioRxiv [Preprint]. 2022 Apr 5:2022.04.05.487060. doi: 10.1101/2022.04.05.487060. bioRxiv. 2022. Update in: Nat Chem Biol. 2023 Mar;19(3):275-283. doi: 10.1038/s41589-022-01149-6. PMID: 35411336 Free PMC article. Updated. Preprint.
References
-
- Lipton SA Paradigm shift in neuroprotection by NMDA receptor blockade: memantine and beyond. Nat. Rev. Drug Discov. 5, 160–170 (2006). - PubMed
References for Methods
-
- Uehara T et al. S-Nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature 441, 513–517 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P41 GM103533/GM/NIGMS NIH HHS/United States
- T32 EB009380/EB/NIBIB NIH HHS/United States
- P20 GM119943/GM/NIGMS NIH HHS/United States
- R61 NS122098/NS/NINDS NIH HHS/United States
- R35 AG071734/AG/NIA NIH HHS/United States
- R01 AG056259/AG/NIA NIH HHS/United States
- T32 AI007384/AI/NIAID NIH HHS/United States
- DP1 DA041722/DA/NIDA NIH HHS/United States
- R01 AG061845/AG/NIA NIH HHS/United States
- UM1 AI144462/AI/NIAID NIH HHS/United States
- R01 DA048882/DA/NIDA NIH HHS/United States
- RF1 AG057409/AG/NIA NIH HHS/United States
- R01 GM132826/GM/NIGMS NIH HHS/United States
- RF1 NS123298/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

