The NALCN channel regulates metastasis and nonmalignant cell dissemination
- PMID: 36175792
- PMCID: PMC9729110
- DOI: 10.1038/s41588-022-01182-0
The NALCN channel regulates metastasis and nonmalignant cell dissemination
Abstract
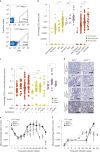
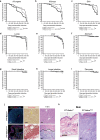
We identify the sodium leak channel non-selective protein (NALCN) as a key regulator of cancer metastasis and nonmalignant cell dissemination. Among 10,022 human cancers, NALCN loss-of-function mutations were enriched in gastric and colorectal cancers. Deletion of Nalcn from gastric, intestinal or pancreatic adenocarcinomas in mice did not alter tumor incidence, but markedly increased the number of circulating tumor cells (CTCs) and metastases. Treatment of these mice with gadolinium-a NALCN channel blocker-similarly increased CTCs and metastases. Deletion of Nalcn from mice that lacked oncogenic mutations and never developed cancer caused shedding of epithelial cells into the blood at levels equivalent to those seen in tumor-bearing animals. These cells trafficked to distant organs to form normal structures including lung epithelium, and kidney glomeruli and tubules. Thus, NALCN regulates cell shedding from solid tissues independent of cancer, divorcing this process from tumorigenesis and unmasking a potential new target for antimetastatic therapies.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures












Comment in
-
Skip to the end: metastasis before tumorigenesis.Nat Genet. 2022 Dec;54(12):1774-1775. doi: 10.1038/s41588-022-01228-3. Nat Genet. 2022. PMID: 36369288 Free PMC article.
-
Uncoupling Metastasis from Tumorigenesis.N Engl J Med. 2023 Feb 16;388(7):657-659. doi: 10.1056/NEJMcibr2213497. N Engl J Med. 2023. PMID: 36791168 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

