Construction of a focal adhesion signaling pathway-related ceRNA network in pelvic organ prolapse by transcriptome analysis
- PMID: 36176289
- PMCID: PMC9513229
- DOI: 10.3389/fgene.2022.996310
Construction of a focal adhesion signaling pathway-related ceRNA network in pelvic organ prolapse by transcriptome analysis
Abstract
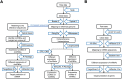
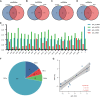
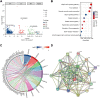
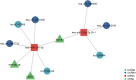
Objective: Pelvic organ prolapse (POP) affects a large proportion of adult women, but the pathogenesis of POP remains unclear. The increase in global population aging will impose a substantial medical burden. Herein, we aimed to explore the related RNAs regulating the occurrence of POP and provide potential therapeutic targets. Method: Tissue biopsies were collected from the anterior vaginal wall of six women with POP and six matched subjects without POP. The profiles of mRNAs, circRNAs, lncRNAs, and miRNAs were obtained by whole transcriptome RNA sequencing. Result: The findings revealed that 71 circRNAs, 76 known lncRNAs, 84 miRNAs, and 931 mRNAs were significantly altered (p < 0.05 and |log2FC| > 1). GO and KEGG enrichment analyses indicated that the differentially expressed genes (DEGs) were mainly enriched in the focal adhesion signaling pathway. FLT, ITGA9, VEGFD, PPP1R12B, and ROCK2 were identified as focal adhesion signaling pathway-related hub genes by protein-protein interaction network analysis. Based on the relationships between the DEGs and miRNA, lncRNA and circRNA targets, we constructed a focal adhesion signaling pathway-related ceRNA network. The ceRNA network includes hsa_circ_0002190/hsa_circ_0046843/lnc-CARMN -miR-23a-3p - ROCK2 and hsa_circ_0001326/hsa_circ_0007733/lnc-AC107959/lnc-TPM1-AS - miR-205-5p - ROCK2/PPP1R12B/VEGFD. Moreover, abnormalities in the cytoskeleton in fibroblasts from individuals with POP were observed. Conclusion: In this study, a focal adhesion signaling pathway-related ceRNA network was constructed, and this network may serve as a target for finding suitable drugs for the treatment of POP.
Keywords: PPI; competing endogenous RNA; focal adhesion signaling pathway; pelvic organ prolapse; whole transcriptome RNA sequencing.
Copyright © 2022 Yu, He, Chen, Lin, Liu, Yang, Ye, Zheng, Yang and Lin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

