Extracellular matrix stiffness controls cardiac valve myofibroblast activation through epigenetic remodeling
- PMID: 36176599
- PMCID: PMC9472021
- DOI: 10.1002/btm2.10394
Extracellular matrix stiffness controls cardiac valve myofibroblast activation through epigenetic remodeling
Abstract
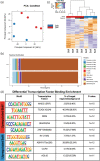
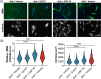
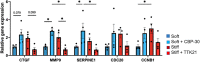
Aortic valve stenosis (AVS) is a progressive fibrotic disease that is caused by thickening and stiffening of valve leaflets. At the cellular level, quiescent valve interstitial cells (qVICs) activate to myofibroblasts (aVICs) that persist within the valve tissue. Given the persistence of myofibroblasts in AVS, epigenetic mechanisms have been implicated. Here, we studied changes that occur in VICs during myofibroblast activation by using a hydrogel matrix to recapitulate different stiffnesses in the valve leaflet during fibrosis. We first compared the chromatin landscape of qVICs cultured on soft hydrogels and aVICs cultured on stiff hydrogels, representing the native and diseased phenotypes respectively. Using assay for transposase-accessible chromatin sequencing (ATAC-Seq), we found that open chromatin regions in aVICs were enriched for transcription factor binding motifs associated with mechanosensing pathways compared to qVICs. Next, we used RNA-Seq to show that the open chromatin regions in aVICs correlated with pro-fibrotic gene expression, as aVICs expressed higher levels of contractile fiber genes, including myofibroblast markers such as alpha smooth muscle actin (αSMA), compared to qVICs. In contrast, chromatin remodeling genes were downregulated in aVICs compared to qVICs, indicating qVICs may be protected from myofibroblast activation through epigenetic mechanisms. Small molecule inhibition of one of these remodelers, CREB Binding Protein (CREBBP), prevented qVICs from activating to aVICs. Notably, CREBBP is more abundant in valves from healthy patients compared to fibrotic valves. Our findings reveal the role of mechanical regulation in chromatin remodeling during VIC activation and quiescence and highlight one potential therapeutic target for treating AVS.
Keywords: ATAC‐Seq; RNA‐Seq; epigenetics; hydrogels; myofibroblasts; valve interstitial cells.
© 2022 The Authors. Bioengineering & Translational Medicine published by Wiley Periodicals LLC on behalf of American Institute of Chemical Engineers.
Conflict of interest statement
The authors declare no conflicts of interests.
Figures





References
-
- Rajamannan NM, Evans FJ, Aikawa E, et al. Calcific aortic valve disease: not simply a degenerative process: a review and agenda for research from the national heart and lung and blood institute aortic stenosis working group. Circulation. 2011;124(16):1783‐1791. doi: 10.1161/CIRCULATIONAHA.110.006767 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

