An integrated bioinformatic investigation of mitochondrial energy metabolism genes in colon adenocarcinoma followed by preliminary validation of CPT2 in tumor immune infiltration
- PMID: 36177002
- PMCID: PMC9513322
- DOI: 10.3389/fimmu.2022.959967
An integrated bioinformatic investigation of mitochondrial energy metabolism genes in colon adenocarcinoma followed by preliminary validation of CPT2 in tumor immune infiltration
Abstract
Background: The prognosis for colon adenocarcinoma (COAD) today remains poor. Changes in mitochondria-related genes and metabolic reprogramming are related to tumor growth, metastasis, and immune evasion and are key factors in tumor genesis and development.
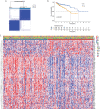
Methods: TCGA database was used to analyze the differentially expressed mitochondrial energy metabolism pathway-related genes (MMRGs) in COAD patients, and the mutation of MMRG in tumor cells, the biological processes involved, and the correlation with tumor immunity were also analyzed. Then, MMRG and MMRG-related genes were used to divide COAD patients into different subtypes, and immunocorrelation analysis and survival analysis were performed. Finally, univariate regression analysis and LASSO regression analysis were used to construct a prognostic risk model for COAD patients, which was verified by the GEO database and evaluated by Kaplan-Meier (K-M) and receiver operating characteristic (ROC) curves, and the correlation between the risk model and immunity and clinical subtypes based on MMRG was analyzed.
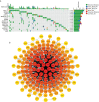
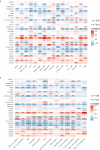
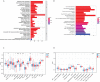
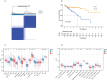
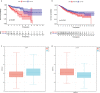
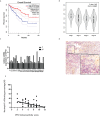
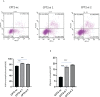
Results: In this study, the MMRG patterns and tumor immune microenvironment characteristics in COAD patients were systematically evaluated by clustering the expression of 188 MMRGs. We identified two subtypes of COAD with different clinical and immunological characteristics. Eight of the 28 differentially expressed MMRG genes were used to construct risk scores. ROC and K-M curves suggested that the risk model could well predict the prognosis of COAD patients, and the risk model was related to immune cell infiltration and immune function.
Conclusions: The two COAD subtypes identified by MMRG are helpful for the clinical differentiation of patients with different prognoses and tumor progressions, and the risk score can assist the clinical evaluation of patient prognosis. Our results suggest that CPT2 contributes to the recruitment and regulation of neutrophils in COAD. CPT2 may act as a valuable biomarker for COAD immunotherapy.
Keywords: CPT2; colon adenocarcinoma; energy metabolism; immune; mitochondrion.
Copyright © 2022 Cao, Lin, Fu, Niu, Yang and Cai.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- de Jong MC, Pulitano C, Ribero D, Strub J, Mentha G, Schulick RD, et al. Rates and patterns of recurrence following curative intent surgery for colorectal liver metastasis: an international multi-institutional analysis of 1669 patients. Ann Surg (2009) 250:440–8. doi: 10.1097/SLA.0b013e3181b4539b - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

