HLAII peptide presentation of infliximab increases when complexed with TNF
- PMID: 36177046
- PMCID: PMC9513746
- DOI: 10.3389/fimmu.2022.932252
HLAII peptide presentation of infliximab increases when complexed with TNF
Abstract
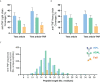
CD4+ T-cell activation through recognition of Human Leukocyte Antigen II (HLAII)-presented peptides is a key step in the development of unwanted immune response against biotherapeutics, such as the generation of anti-drug antibodies (ADA). Therefore, the identification of HLAII-presented peptides derived from biotherapeutics is a crucial part of immunogenicity risk assessment and mitigation strategies during drug development. To date, numerous CD4+ T-cell epitopes have been identified by HLAII immunopeptidomics in antibody-based biotherapeutics using either their native or aggregated form. Antibody-target immune complexes have been detected in patients with ADA and are thought to play a role in ADA development by enhancing the presentation of CD4+ T-cell epitopes at the surface of antigen presenting cells (APCs). The aim of this study was to investigate the effect of biotherapeutic antibody-target immune complexes on the HLAII peptide presentation of biotherapeutics in human primary monocyte-derived dendritic cells (DCs). The trimeric tumor necrosis factor (TNF) and its biotherapeutic antagonists infliximab (INFL), adalimumab (ADAL), and a single armed Fab' were used as a model system. The HLAII immunopeptidome of DCs loaded with antagonists or their immune complexes with TNF was analyzed by trapped ion mobility time-of-flight mass spectrometry (timsTOF MS) leading to the identification of ~ 12,000 unique HLAII-associated peptides per preparation. Anti-TNF sequences were detected at a median of 0.3% of the total immunopeptidome, against a majority background of peptides from endogenous and media-derived proteins. TNF antagonist presentation spanned the variable and constant regions in a widespread manner in both light and heavy chains, consistent with previously discovered HLAII peptides. This investigation extends the collection of observed HLAII peptides from anti-TNF biotherapeutics to include sequences that at least partially span the complementary determining regions (CDRs), such as the LCDR1 for both INFL and ADAL. Although antagonist presentation varied significantly across donors, peptides from both bivalent antagonists INFL and ADAL were more highly presented relative to the Fab'. While TNF immune complexes did not alter overall HLAII presentation, a moderate increase in presentation of a subset of peptide clusters was observed in the case of INFL-TNF, which included HCDR2, HCDR3 and LCDR2 sequences.
Keywords: CD4+ T-cells; HLAII; TNF antagonist; dendritic cells; immunogenicity; immunopeptidomics; presented peptides; timsTOF mass spectrometry.
Copyright © 2022 Casasola-LaMacchia, Seward, Tourdot, Willetts, Kruppa, Agostino, Bergeron, Ahyi-Amendah, Ciarla, Lu, Kim, Hickling and Neubert.
Conflict of interest statement
AC, GB, ZL, H-YK, HN, RS, ST, and AC-L are current employees of Pfizer Inc. MA, NA-A, and TH were employed at Pfizer when they contributed to this study. GK and MW are current employees of Bruker Daltonics.
Figures


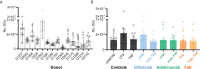



Similar articles
-
Secukinumab, a novel anti-IL-17A antibody, shows low immunogenicity potential in human in vitro assays comparable to other marketed biotherapeutics with low clinical immunogenicity.MAbs. 2016;8(3):536-50. doi: 10.1080/19420862.2015.1136761. Epub 2016 Jan 28. MAbs. 2016. PMID: 26817498 Free PMC article.
-
Development of a semi-automated MHC-associated peptide proteomics (MAPPs) method using streptavidin bead-based immunoaffinity capture and nano LC-MS/MS to support immunogenicity risk assessment in drug development.Front Immunol. 2023 Nov 10;14:1295285. doi: 10.3389/fimmu.2023.1295285. eCollection 2023. Front Immunol. 2023. PMID: 38022649 Free PMC article.
-
Internalization of therapeutic antibodies into dendritic cells as a risk factor for immunogenicity.Front Immunol. 2024 Aug 28;15:1406643. doi: 10.3389/fimmu.2024.1406643. eCollection 2024. Front Immunol. 2024. PMID: 39263220 Free PMC article.
-
Immunogenicity of TNF-Inhibitors.Front Immunol. 2020 Feb 26;11:312. doi: 10.3389/fimmu.2020.00312. eCollection 2020. Front Immunol. 2020. PMID: 32174918 Free PMC article. Review.
-
Hunting down factor VIII in the immunopeptidome.Cell Immunol. 2016 Mar;301:59-64. doi: 10.1016/j.cellimm.2015.11.001. Epub 2015 Nov 5. Cell Immunol. 2016. PMID: 26610639 Review.
Cited by
-
Immunogenicity risk assessment and mitigation for engineered antibody and protein therapeutics.Nat Rev Drug Discov. 2024 Dec;23(12):898-913. doi: 10.1038/s41573-024-01051-x. Epub 2024 Oct 18. Nat Rev Drug Discov. 2024. PMID: 39424922 Review.
-
Antidrug antibodies to adalimumab do not associate with immunologically related adverse events.Front Immunol. 2025 Feb 27;15:1457993. doi: 10.3389/fimmu.2024.1457993. eCollection 2024. Front Immunol. 2025. PMID: 40084239 Free PMC article. Clinical Trial.
-
HLAIIPred: cross-attention mechanism for modeling the interaction of HLA class II molecules with peptides.Commun Biol. 2025 Jul 30;8(1):1133. doi: 10.1038/s42003-025-08500-2. Commun Biol. 2025. PMID: 40739437 Free PMC article.
-
Sensitive, High-Throughput HLA-I and HLA-II Immunopeptidomics Using Parallel Accumulation-Serial Fragmentation Mass Spectrometry.Mol Cell Proteomics. 2023 Jun;22(6):100563. doi: 10.1016/j.mcpro.2023.100563. Epub 2023 May 3. Mol Cell Proteomics. 2023. PMID: 37142057 Free PMC article.
-
TOFIMS mass spectrometry-based immunopeptidomics refines tumor antigen identification.Nat Commun. 2023 Nov 17;14(1):7472. doi: 10.1038/s41467-023-42692-7. Nat Commun. 2023. PMID: 37978195 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

