GeneNetTools: tests for Gaussian graphical models with shrinkage
- PMID: 36179082
- PMCID: PMC9665865
- DOI: 10.1093/bioinformatics/btac657
GeneNetTools: tests for Gaussian graphical models with shrinkage
Abstract
Motivation: Gaussian graphical models (GGMs) are network representations of random variables (as nodes) and their partial correlations (as edges). GGMs overcome the challenges of high-dimensional data analysis by using shrinkage methodologies. Therefore, they have become useful to reconstruct gene regulatory networks from gene-expression profiles. However, it is often ignored that the partial correlations are 'shrunk' and that they cannot be compared/assessed directly. Therefore, accurate (differential) network analyses need to account for the number of variables, the sample size, and also the shrinkage value, otherwise, the analysis and its biological interpretation would turn biased. To date, there are no appropriate methods to account for these factors and address these issues.
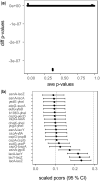
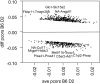
Results: We derive the statistical properties of the partial correlation obtained with the Ledoit-Wolf shrinkage. Our result provides a toolbox for (differential) network analyses as (i) confidence intervals, (ii) a test for zero partial correlation (null-effects) and (iii) a test to compare partial correlations. Our novel (parametric) methods account for the number of variables, the sample size and the shrinkage values. Additionally, they are computationally fast, simple to implement and require only basic statistical knowledge. Our simulations show that the novel tests perform better than DiffNetFDR-a recently published alternative-in terms of the trade-off between true and false positives. The methods are demonstrated on synthetic data and two gene-expression datasets from Escherichia coli and Mus musculus.
Availability and implementation: The R package with the methods and the R script with the analysis are available in https://github.com/V-Bernal/GeneNetTools.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2022. Published by Oxford University Press.
Figures

References
-
- Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc., 57, 289–300.

