Leveraging weak supervision to perform named entity recognition in electronic health records progress notes to identify the ophthalmology exam
- PMID: 36179600
- PMCID: PMC9901505
- DOI: 10.1016/j.ijmedinf.2022.104864
Leveraging weak supervision to perform named entity recognition in electronic health records progress notes to identify the ophthalmology exam
Abstract
Objective: To develop deep learning models to recognize ophthalmic examination components from clinical notes in electronic health records (EHR) using a weak supervision approach.
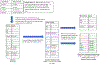
Methods: A corpus of 39,099 ophthalmology notes weakly labeled for 24 examination entities was assembled from the EHR of one academic center. Four pre-trained transformer-based language models (DistilBert, BioBert, BlueBert, and ClinicalBert) were fine-tuned to this named entity recognition task and compared to a baseline regular expression model. Models were evaluated on the weakly labeled test dataset, a human-labeled sample of that set, and a human-labeled independent dataset.
Results: On the weakly labeled test set, all transformer-based models had recall > 0.93, with precision varying from 0.815 to 0.843. The baseline model had lower recall (0.769) and precision (0.682). On the human-annotated sample, the baseline model had high recall (0.962, 95 % CI 0.955-0.067) with variable precision across entities (0.081-0.999). Bert models had recall ranging from 0.771 to 0.831, and precision >=0.973. On the independent dataset, precision was 0.926 and recall 0.458 for BlueBert. The baseline model had better recall (0.708, 95 % CI 0.674-0.738) but worse precision (0.399, 95 % CI -0.352-0.451).
Conclusion: We developed the first deep learning system to recognize eye examination components from clinical notes, leveraging a novel opportunity for weak supervision. Transformer-based models had high precision on human-annotated labels, whereas the baseline model had poor precision but higher recall. This system may be used to improve cohort and feature identification using free-text notes.Our weakly supervised approach may help amass large datasets of domain-specific entities from EHRs in many fields.
Keywords: Deep learning; Electronic health records; Named entity recognition; Natural language processing; Ophthalmology; Weak supervision.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Vaswani A, Shazeer N, Parmar N, et al. Attention Is All You Need. arXiv [cs.CL]. 2017.http://arxiv.org/abs/1706.03762
-
- Huang K, Altosaar J, Ranganath R. ClinicalBERT: Modeling Clinical Notes and Predicting Hospital Readmission. arXiv [cs.CL]. 2019.http://arxiv.org/abs/1904.05342
-
- Alsentzer E, Murphy JR, Boag W, et al. Publicly Available Clinical BERT Embeddings. arXiv [cs.CL]. 2019.http://arxiv.org/abs/1904.03323
-
- Peng Y, Yan S, Lu Z. Transfer Learning in Biomedical Natural Language Processing: An Evaluation of BERT and ELMo on Ten Benchmarking Datasets. arXiv [cs.CL]. 2019.http://arxiv.org/abs/1906.05474
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

