ANGEL2 phosphatase activity is required for non-canonical mitochondrial RNA processing
- PMID: 36180430
- PMCID: PMC9525292
- DOI: 10.1038/s41467-022-33368-9
ANGEL2 phosphatase activity is required for non-canonical mitochondrial RNA processing
Abstract
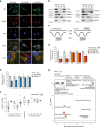
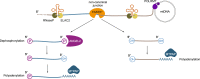
Canonical RNA processing in mammalian mitochondria is defined by tRNAs acting as recognition sites for nucleases to release flanking transcripts. The relevant factors, their structures, and mechanism are well described, but not all mitochondrial transcripts are punctuated by tRNAs, and their mode of processing has remained unsolved. Using Drosophila and mouse models, we demonstrate that non-canonical processing results in the formation of 3' phosphates, and that phosphatase activity by the carbon catabolite repressor 4 domain-containing family member ANGEL2 is required for their hydrolysis. Furthermore, our data suggest that members of the FAST kinase domain-containing protein family are responsible for these 3' phosphates. Our results therefore propose a mechanism for non-canonical RNA processing in metazoan mitochondria, by identifying the role of ANGEL2.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

